31 Bacterial Lysis and Genomic DNA Isolation
Growth of Isolates
- A colony of each of your isolates will be used to start a liquid culture in Tryptic Soy Broth (TSB).
- Grow liquid cultures with shaking at 30°C for 16-18 hours.
Bacterial Lysis
- For general bacterial inputs, reconstitute the lysozyme in TE buffer to 10 mg/ml. We recommend preparing enzymes freshly on the day as enzymatic activity reduces over time once reconstituted.
- In a fresh 1.5 ml Eppendorf DNA LoBind tube, centrifuge 1ml (~1 x 10^8 – 10^9 cfu/ml) of cell culture or 1/8 of a 10 µl loop of colonies from a plate at 12,000 x g for 1 minute.
- Remove the supernatant by pipetting and resuspend the pellet in 1 ml of PBS.
- Centrifuge at 12,000 x g for 1 minute.
- Depending on your input, remove the supernatant and resuspend as follows:
a. For most bacterial inputs: Remove the supernatant and resuspend the pellet in 100 µl TE buffer with 10 µl lysozyme.
b. For staphylococcal inputs: Remove the supernatant and resuspend the pellet in 100 µl Staphylococcal Lysis Buffer (SLB) with 10 µl achromopeptidase. Note: It is important to remove as much PBS as possible without disturbing the pellet as the buffer has an inhibitory effect on achromopeptidase activity.
6. Incubate at 37°C for 10 minutes in a thermomixer at 500 RPM. If a thermomixer is unavailable, the reaction can be incubated stationary and agitated by gently flicking for 10 seconds every two minutes.
7.Combine the following reagents with your sample in the 1.5 ml Eppendorf DNA LoBind tube, and mix by vortexing.
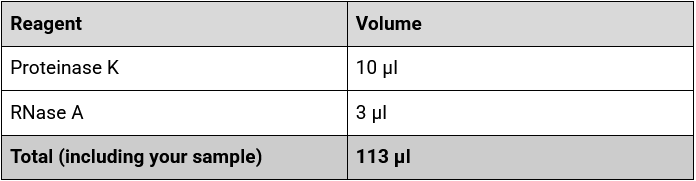
8. Vortex mixing your sample before adding the CTAB buffer allows the enzymes to fully mix with the bacterial cells as the mixture can become viscous.
9. Add 200 µl of CTAB buffer and mix by vortexing.
10. Extractions performed using the CTAB buffer improves sequencing health and output compared to using NEB Lysis Buffer from the Monarch Spin gDNA Extraction Kit. IMPORTANT: The full 30-minute incubation must be completed to ensure the proteinase and RNase activity occurs, regardless of turbidity.
11. Incubate at 56°C for 30 minutes in a thermomixer at 1000 RPM. Note: Depending on the bacterial input, the solution may become fully transparent or may remain hazy. INFO: The Monarch Spin gDNA Extraction Kit is used for gDNA extraction. The remainder of this method is written following the Genomic DNA Binding and Elution protocol with our recommended alterations.
12. Preheat the Monarch® gDNA Elution Buffer to 60°C for later use. IMPORTANT: The ratio of binding buffer to lysate (2:1) is important for the column binding efficiency.
13. We recommend taking through 200 µl of lysate for use with 400 µl of binding buffer per sample.
14. To run more samples from the same isolate, the volumes can be increased in relation to the 2:1 ratio of buffer to lysate.
15. Combine the following reagents in a clean 1.5 ml Eppendorf DNA LoBind tube:
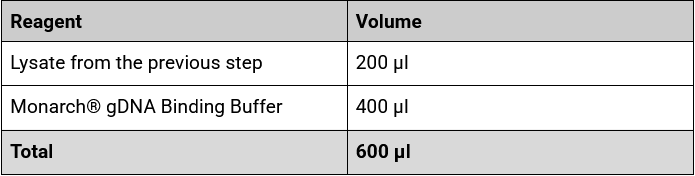
16. Ensure the reaction is thoroughly mixed by pipetting gently until the liquid is homogenous.
gDNA Binding and Elution
17. Thorough mixing is essential to ensure the DNA binds properly to the column and to homogenise the CTAB and Monarch® gDNA Binding Buffer which solidify on initial contact.
18. Transfer the reaction to a Monarch® Spin Column pre-inserted into a Monarch® collection tube, without touching the upper column area.
19. Close the tube and centrifuge for 3 minutes at 1,000 x g to bind the gDNA and then for 1 minute at maximum speed (>12,000 x g) to clear the membrane.
20. Do not empty or remove the collection tube from the centrifuge when changing settings.
21. Remove the gDNA spin column from the collection tube and discard the tube with the flow-through.
22. Transfer the gDNA spin column to a new collection tube and add 500 µl Monarch® gDNA Wash Buffer. IMPORTANT: Do NOT vortex.
23. Close the collection tube and invert multiple times until the wash buffer reaches the cap.
24. Immediately centrifuge the collection tube for 1 minute at maximum speed (>12,000 x g). Remove the gDNA spin column from the collection tube to discard the flow-through.
25. Re-insert the gDNA spin column into the collection tube and add 500 µl gDNA wash buffer and close the tube.
26. Immediately centrifuge the collection tube for 1 minute at maximum speed (>12,000 x g). Remove the gDNA spin column from the collection tube to discard the flow-through.
27. After centrifugation, ensure there is no liquid in the gDNA spin column above or below the silica membrane. If there is liquid, repeat the centrifugation until all liquid is removed. IMPORTANT: Preheating the Monarch® gDNA Elution Buffer to 60°C is essential for maximum recovery of larger gDNA fragments.
28. Place the gDNA spin column in a DNase-free 1.5 ml microfuge tube and add 100 µl of preheated Monarch® gDNA Elution Buffer. Close the tube and incubate at room temperature for 1 minute. TIP: If extractions are not yielding high enough concentrations of DNA for 200 ng per sample, less gDNA elution buffer can be run through the column to increase the concentration. Note: Elution of less than 100 µl results in lower total yield and elution of less than 50 µl is not recommended.
29. Centrifuge for 1 minute at maximum speed (>12,000 x g) to elute the gDNA into the 1.5 ml microfuge tube.
30. The gDNA spin column can be discarded.
31. CHECKPOINT: Quantify 1 µl of eluted sample using a Qubit fluorometer. Expected yield is ~15-20 ng/µl per sample with a DNA Integrity Number (DIN) of 9.
32. END OF STEP: Take forward 200 ng per sample into the library preparation step of the Nanopore-only Microbial Isolate Sequencing Solution (NO-MISS) – Rapid Barcoding Kit V14 (SQK-RBK114.24 or SQK-RBK114.96) end-to-end protocol.
We recommend storing the DNA frozen (-30 to -15°C or -90 to -65°C). We will freeze the DNA and use for quantification in the following lab sessions before sequencing.

