Enzymes are so large, so structurally complex, and so numerous that the use of computer databases and molecular visualization programs has become an essential tool for studying biological chemistry. Of the various databases available online, the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (http://www.genome.jp/kegg/pathway.html), maintained by the Kanehisa Laboratory of Kyoto University Bioinformatics Center, is useful for obtaining information on biosynthetic pathways of the sort we’ll be describing in Chapter 29. For obtaining information on a specific enzyme, the BRENDA database (http://www.brenda-enzymes.org), maintained by the Institute of Biochemistry at the University of Cologne, Germany, is particularly valuable.
Perhaps the most useful of all biological databases is the Protein Data Bank (PDB), operated by the Research Collaboratory for Structural Bioinformatics (RCSB). The PDB is a worldwide repository of X-ray and NMR structural data for biological macromolecules. In mid-2022, data for more than 192,888 structures were available, and more than 7000 new ones were being added yearly. To access the Protein Data Bank, go to www.rcsb.org and a home page like that shown in Figure 26.12 will appear. As with much that is available online, however, the PDB site is changing rapidly, so you may not see quite the same thing.
Figure 26.12 The Protein Data Bank home page. (credit: RCSB Protein Data Bank)
To learn how to use the PDB, begin by running the short tutorial listed under Learn on the left side column. After that introduction, start exploring. Let’s say you want to view citrate synthase, the enzyme that catalyzes the addition of acetyl CoA to oxaloacetate to give citrate. Type “citrate synthase” (with quotation marks) into the small search box on the top line, click on “Search,” and a list of 2,400 or so structures will appear. Scroll down near the end of the list until you find the entry with a PDB code of 5CTS and the title “Proposed Mechanism for the Condensation Reaction of Citrate Synthase: 1.9-Angstroms Structure of the Ternary Complex with Oxaloacetate and Carboxymethyl Coenzyme A.” Alternatively, if you know the code of the enzyme you want, you can enter it directly into the search box.
Click on the PDB code of entry 5CTS, and a new page containing information about the enzyme will open.
If you choose, you can download the structure file to your computer and open it with any of numerous molecular graphics programs to see an image like that in Figure 26.13. The biologically active molecule is a dimer of two identical subunits consisting primarily of α– helical regions displayed as coiled ribbons. For now, just select “View in Jmol” under the enzyme image on the right side of the screen to see some of the options for visualizing and further exploring the enzyme.
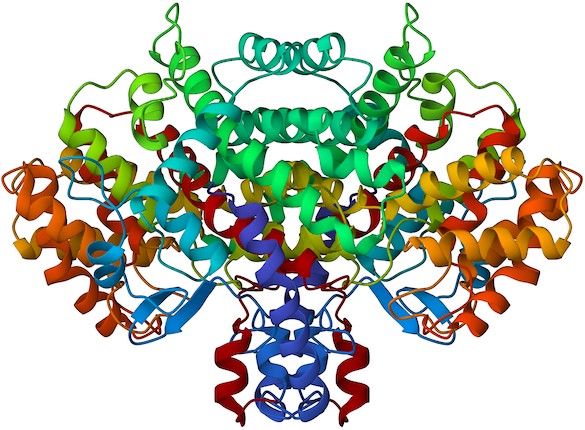
Figure 26.13 An image of citrate synthase, downloaded from the Protein Data Bank. (credit: Protein Data Bank, 1AL6 B. PDB ID: 1AL6 B. Schwartz, K.W. Vogel, K.C. Usher, C. Narasimhan, H.M. Miziorko, S.J. Remington, D.G. Drueckhammer. Mechanisms of Enzyme- Catalyzed Deprotonation of Acetyl-Coenzyme A, CC BY 1.0)

