28 Chapter 28 – Biomolecules: Nucleic Acids Solutions to Problems
Facilitators:
28.1
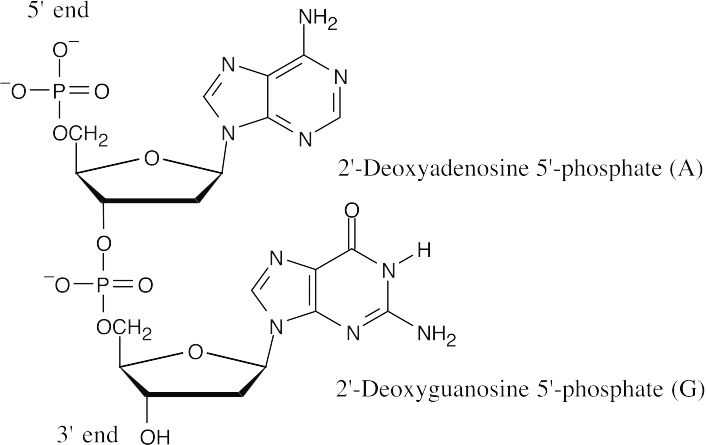
28.2
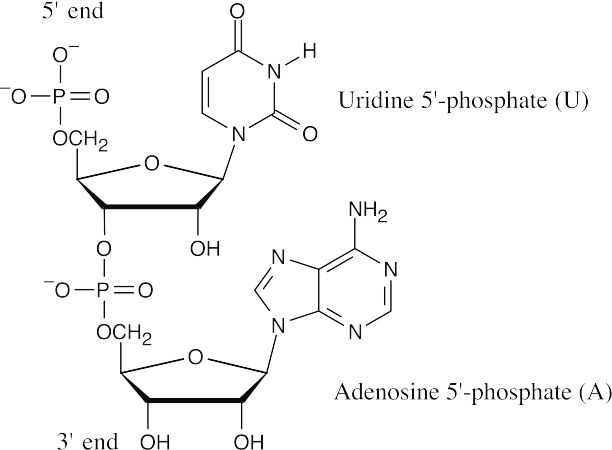
28.3DNA (5′ end) GGCTAATCCGT (3′ end) is complementary .to DNA (3′ end) CCGATTAGGCA (5′ end).
Remember that the complementary strand has the 3′ end on the left and the 5′ end on the right.
The complementary sequence can also be written as:
DNA (5′ end) ACGGATTAGCC (3′ end). Be sure that you know which format is being used (3′ to 5′, or 5′ to 3′).
28.4
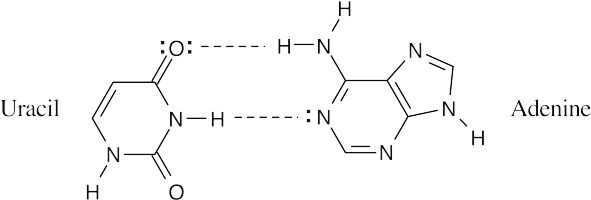
- DNA (5′ end) GATTACCGTA(3′ end) is complementary to RNA (3′ end) CUAAUGGCAU (5′ end)
- RNA (5′ end) UUCGCAGAGU (3′ end)
DNA (3′ end) AAGCGTCTCA (5′ end) antisense (noncoding) strand
28.7–28.8
Several different codons can code for the same amino acid (Table 28.1). The corresponding anticodon follows the slash mark after each codon. The mRNA codons are written with the 5′ end on the left and the 3′ end on the right, and the tRNA anticodons have the 3′ end on the left and the 5′ end on the right.
|
Amino acid: |
Ala |
Phe |
Leu |
Tyr |
|
Codon sequence/ |
GCU/CGA |
UUU/AAA |
UUA/AAU |
UAU/AUA |
|
tRNA anticodon: |
GCC/CGG |
UUC/AAG |
UUG/AAC |
UAC/AUG |
|
|
GCA/CGU |
|
CUU/GAA |
|
|
|
GCG/CGC |
|
CUC/GAG |
|
|
|
|
|
CUA/GAU |
|
|
|
|
|
CUG/GAC |
|
28.9–28.10
The mRNA base sequence:(5′ end)CUU–AUG–GCU–UGG–CCC–UAA (3′ end) The amino acid sequence:Leu—Met—Ala—Trp—Pro–(stop)
The DNA sequence: (antisense strand)
(3′ end)GAA–TAC–CGA–ACC–GGG–ATT (5′ end)
28.11
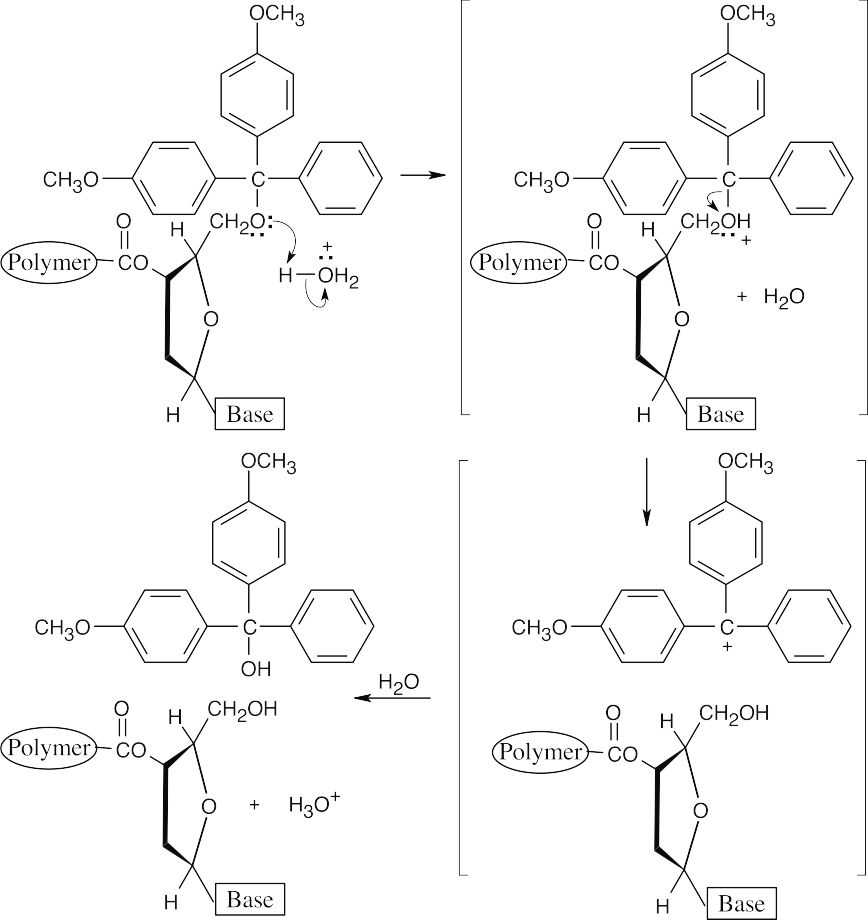
Cleavage of DMT ethers proceeds by an SN1 mechanism and is rapid because the DMT cation is unusually stable.
28.12
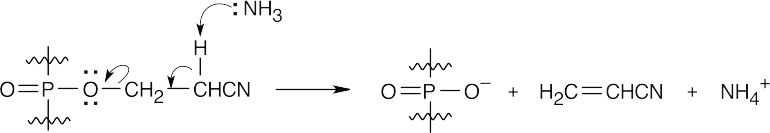
This is an E2 elimination reaction, which proceeds easily because the hydrogen α to the nitrile group is acidic.
Additional Problems
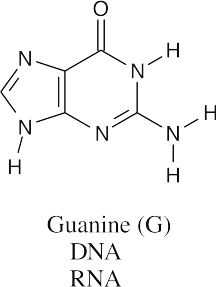 Visualizing Chemistry 28.13
Visualizing Chemistry 28.13
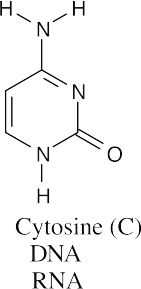
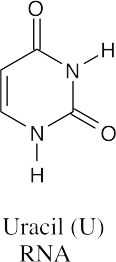 (b)(c)
(b)(c)
All three bases are found in RNA, but only guanine and cytosine are found in DNA.
28.14
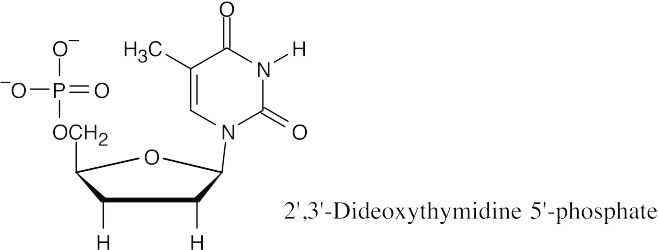
The triphosphate made from 2′,3′-dideoxythymidine 5′ phosphate is labeled with a fluorescent dye and used in the Sanger method of DNA sequencing. Along with the restriction fragment to be sequenced, a DNA primer, and a mixture of the four dNTPs, small quantities of the four labeled dideoxyribonucleotide triphosphates are mixed together. DNA polymerase is added, and a strand of DNA complementary to the restriction fragment is synthesized. Whenever a dideoxyribonucleotide is incorporated into the DNA chain, chain growth stops. The fragments are separated by electrophoresis, and each terminal dideoxynucleotide can be identified by the color of its fluorescence. By identifying these terminal dideoxynucleotides, the sequence of the restriction fragment can be read.
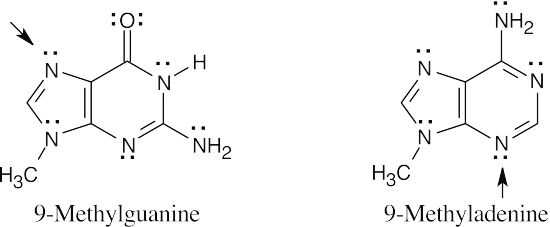
- According to the electrostatic potential map, the nitrogen at the 7 position of 9- methylguanine is more electron-rich (red) and should be more nucleophilic. Thus 9- methylguanine should be the better nucleophile.
Mechanism Problems
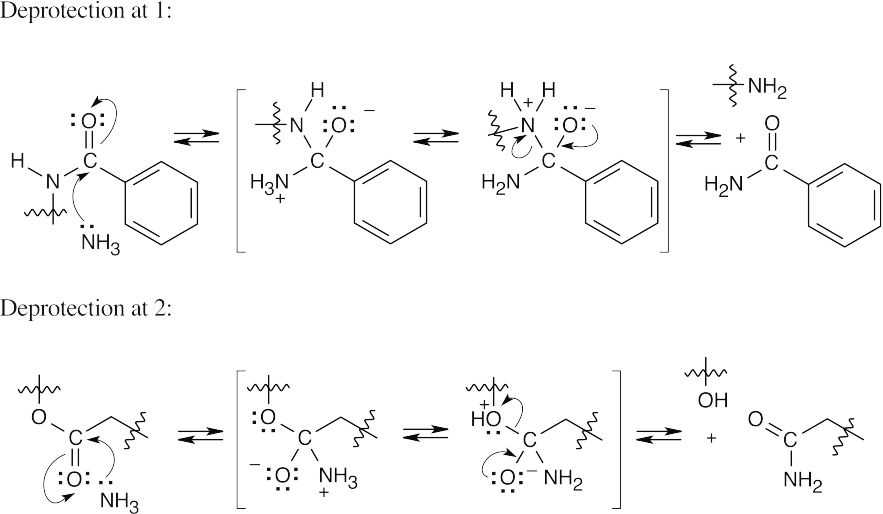
- Both of these cleavages occur by the now-familiar nucleophilic acyl substitution route. A nucleophile adds to the carbonyl group, a proton shifts location, and a second group is eliminated. Only the reacting parts of the structures are shown.
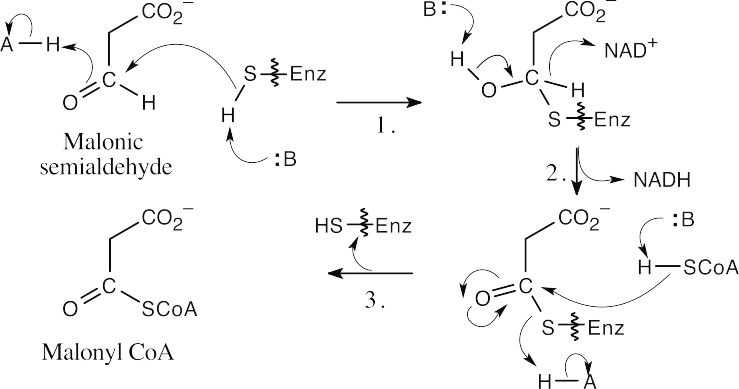
- This reaction involves addition of a thiol residue of the enzyme to malonic semialdehyde, yielding a hemithioacetal (Step 1). Oxidation by NAD+ (step 2), followed by nucleophilic acyl substitution by CoA (Step 3), gives malonyl CoA.
- The steps: (1) phosphorylation by ATP; (2) cyclization; (3) loss of phosphate; (4) tautomerization.
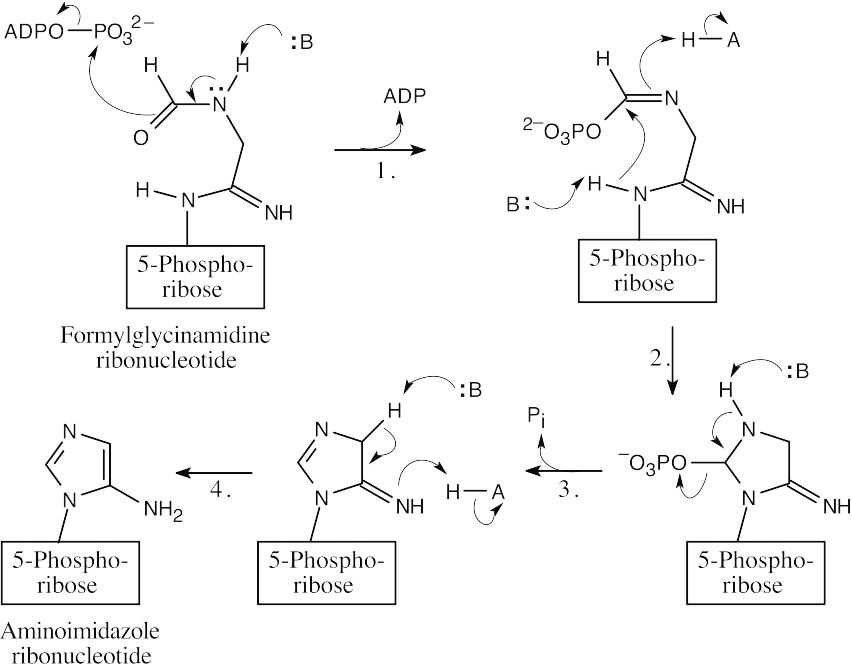
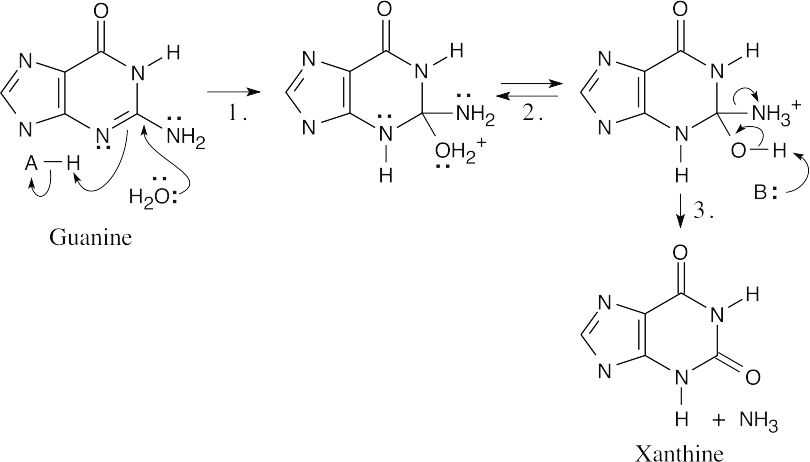
- The steps: (1) Addition of water; (2) Proton shift; (3) Elimination of NH3.
- Both steps are nucleophilic acyl substitutions. (a)
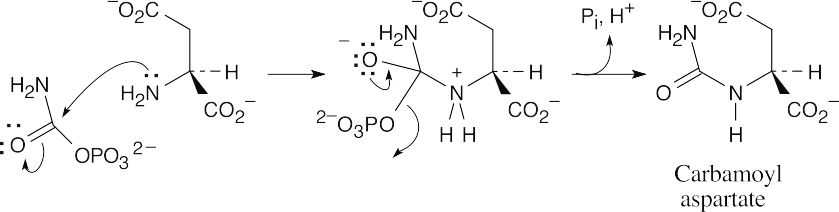
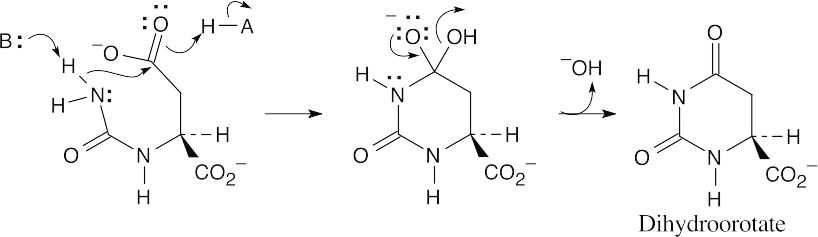 (b)
(b)
General Problems
- The DNA that codes for natriuretic peptide (32 amino acids) consists of 99 bases; 3 bases code for each of the 32 amino acids in the chain (96 bases), and a 3-base “stop” codon is also needed.
Position 9:
Horse amino acid = GlyHuman amino acid = Ser mRNA codons (5′ → 3′):
GGU GGC GGA GGGUCU UCC UCA UCG AGU AGC
DNA bases (antisense strand 3′ → 5′):
CCA CCG CCT CCCAGA AGG AGT AGC TCA TCG
The underlined horse DNA base triplets differ from their human counterparts (also underlined) by only one base.
Position 30:
Horse amino acid = AlaHuman amino acid = Thr mRNA codons (5′ → 3′):
GCU GCC GCA GCGACU ACC ACA ACG
DNA bases (antisense strand 3′ → 5′):
CGA CGG CGT CGCTGA TGG TGT TGC
Each of the above groups of DNA bases from horse insulin has a counterpart in human insulin that differs from it by only one base. It is possible that horse insulin DNA differs from human insulin DNA by only two bases out of 159!
- The percent of A always equals the percent of T, since A and T are complementary. The percent G equals the percent C for the same reason. Thus, sea urchin DNA contains about 32% each of A and T, and about 18% each of G and C.
- Even though the stretch of DNA shown contains UAA in sequence, protein synthesis doesn’t stop. The codons are read as 3-base individual units from start to end, and, in this mRNA sequence, the unit UAA is read as part of two codons, not as a single codon.
- Restriction endonucleases cleave DNA base sequences that are palindromes, meaning that the sequence reads the same as the complement when both are read in the (5′) to (3′) direction. Thus, the sequence in (c), CTCGAG is recognized. The sequence in (a), GAATTC, is also a palindrome and is recognized by a restriction endonuclease. The sequence in (b) is not a palindrome and is not recognized.
28.26–28.28
|
mRNA codon : |
(5’→3′) |
(a) AAU |
(b) GAG |
(c) UCC |
(d) CAU |
|
Amino acid: |
|
Asn |
Glu |
Ser |
His |
|
DNA sequence: |
(3’→5′) |
TTA |
CTC |
AGG |
GTA |
|
tRNA anticodon: |
(3’→5′) |
UUA |
CUC |
AGG |
GUA |
The DNA sequence of the antisense (noncoding) strand is shown.
28.29, 28.30
UAC is a codon for tyrosine. It was transcribed from ATG of the antisense strand of a DNA chain.
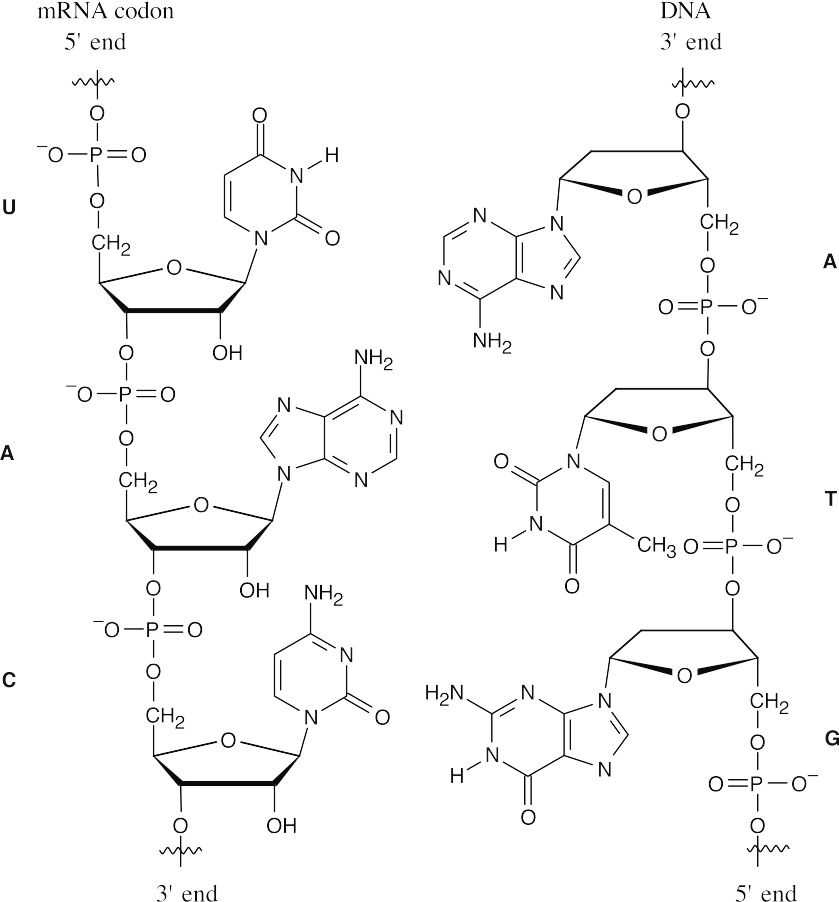
28.31
Tyr———Gly———Gly———Phe———Met(stop)is coded by
|
UAC |
GGU |
GGU |
UUU |
AUG |
UAA |
|
UAU |
GGC |
GGC |
UUC |
|
UAG |
|
|
GGA |
GGA |
|
|
UGA |
|
|
GGG |
GGG |
|
|
|
A total of 2 x 4 x 4 x 2 x 1 x 3 = 194 different mRNA sequences can code for metenkephalin!
- Angiotensin II: Asp—–Arg—–Val—–Tyr—–Ile—–His—–Pro—–Phe (stop)
|
mRNA |
GAU |
CGU |
GUU |
UAU |
AUU |
CAU |
CCU |
UUU |
UAA |
|
sequence: |
|
|
|
|
|
|
|
|
|
|
(5’→3′) |
GAC |
CGC |
GUC |
UAC |
AUC |
CAC |
CCC |
UUC |
UAG |
|
|
CGA |
|
GUA |
|
AUA |
|
CCA |
|
UGA |
|
|
CGG |
|
GUG |
|
|
|
CCG |
|
|
|
|
AGA |
|
|
|
|
|
|
|
|
|
|
AGG |
|
|
|
|
|
|
|
|
As in the previous problem, many mRNA sequences (13,824) can code for angiotensin II.
- DNA coding strand (5’→3′):CTT— CGA—CCA— GAC—AGC—TTT mRNA (5’→3′):CUU—CGA—CCA—GAC—AGC—UUU Amino acid sequence:Leu––––Arg–––Pro–––Asp–––Ser––––Phe
The mRNA sequence is the complement of the DNA noncoding (antisense) strand, which is the complement of the DNA coding (sense) strand. Thus, the mRNA sequence is a copy of the DNA coding (sense) strand, with T replaced by U.
- mRNA sequence (5’→3′): CUA—GAC—CGU—UCC—AAG—UGA Amino Acid:Leu––––Asp–––Arg–––Ser–––Lys(stop)
|
|
Original Sequence |
Miscopied Sequence |
|
DNA coding strand (5’→3′): |
-CAA-CCG-GAT- |
-CGA-CCG-GAT- |
|
mRNA sequence (5’→3′): |
-CAA-CCG-GAU- |
-CGA-CCG-GAU- |
|
Amino acid sequence: |
-Gln––Pro––Asp- |
-Arg––Pro––Asp- |
If this gene sequence were miscopied in the indicated way, a glutamine in the original protein would be replaced by an arginine in the mutated protein.
- 1.First, protect the nucleosides.
- Bases are protected by amide formation.
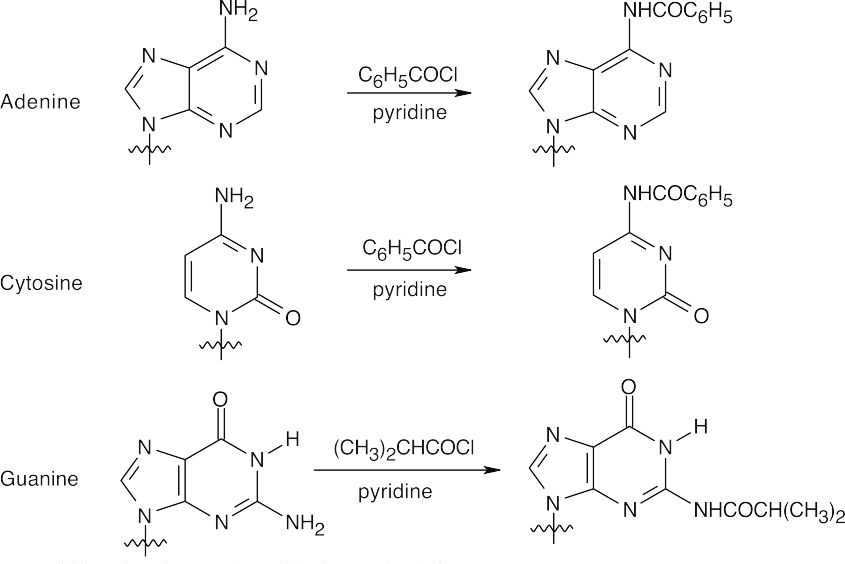
Thymine does not need to be protected.
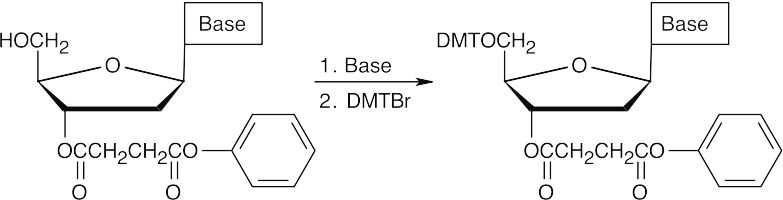
- The 5′ hydroxyl group is protected as its p-dimethoxytrityl (DMT) ether.
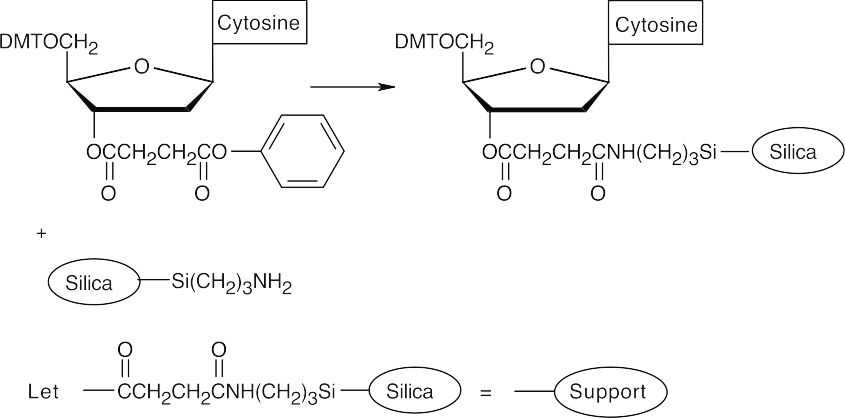
- Attach a protected 2-deoxycytidine nucleoside to the polymer support.
- Cleave the DMT ether.
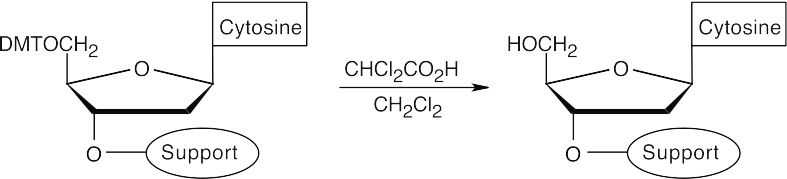
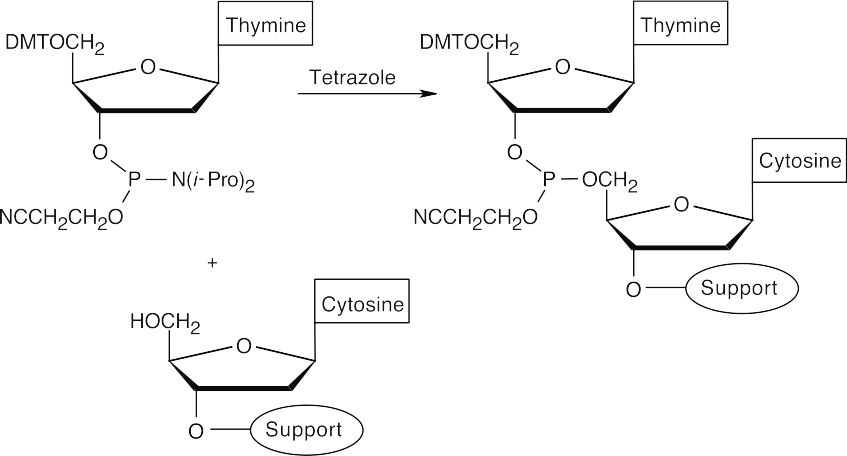
- Couple protected 2′-deoxythymidine to the polymer-2′-deoxycytidine. (The nucleosides have a phosphoramidite group at the 3′ position.)
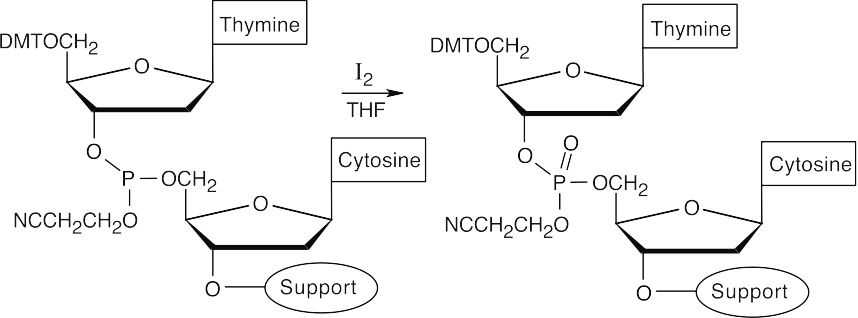
- Oxidize the phosphite product to a phosphate triester, using iodine.
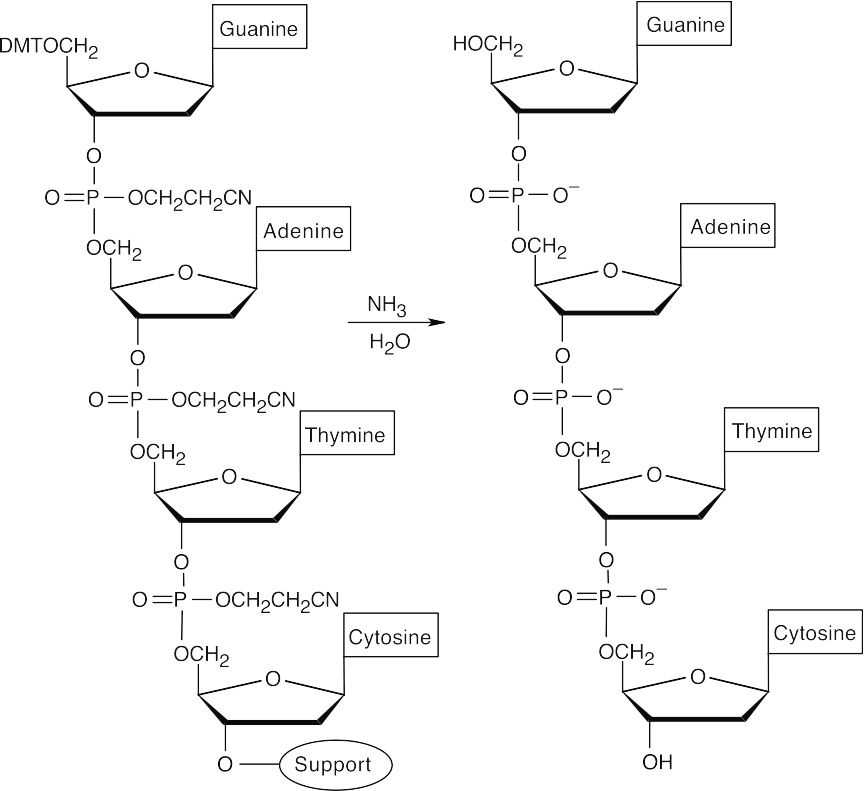
- Repeat steps 3–5 with protected 2′-deoxyadenosine and protected 2′- deoxyguanosine.
- Cleave all protecting groups with aqueous ammonia to yield the desired sequence.
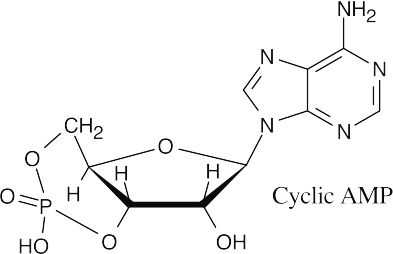
28.38 (a), (b)
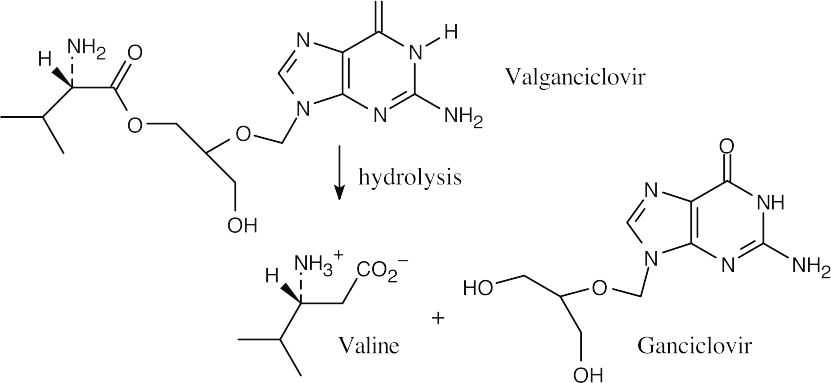
- (c)
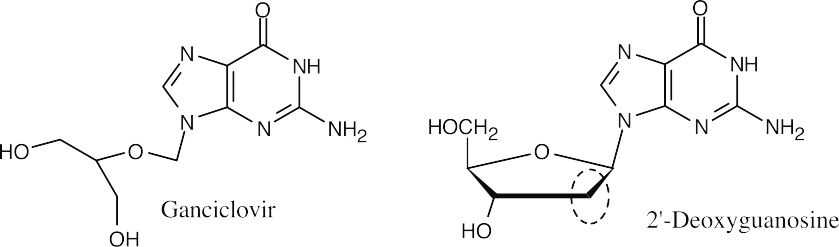
The –CH2– group at C2 of deoxyribose is missing from ganciclovir.
- The missing atoms are part of the relatively inflexible deoxyribose ring. Without the ring, the DNA chain is floppy and base pairing to form a double helix can’t occur.
- As mentioned in (d), the inability to form base pairs stops the replication of DNA.
This file is copyright 2023, Rice University. All Rights Reserved.

