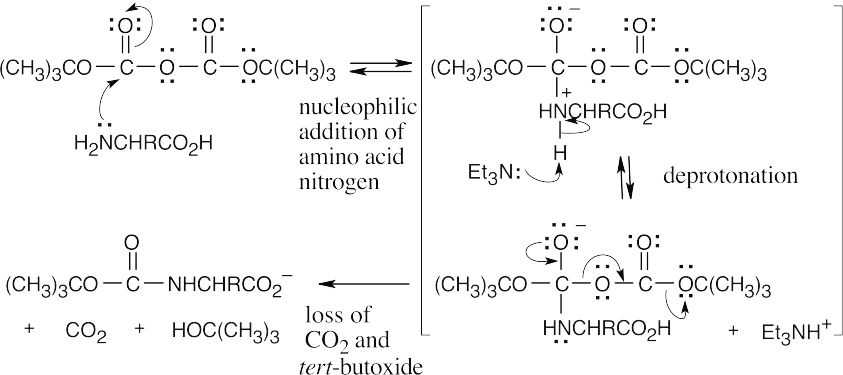26 Chapter 26 – Biomolecules: Amino Acids, Peptides and Proteins Solutions to Problems
26.1Amino Acids with aromatic rings: Phe, Tyr, Trp, His. Amino acids containing sulfur: Cys, Met.
Amino acids that are alcohols: Ser, Thr (Tyr is a phenol.)
Amino acids having hydrocarbon side chains: Ala, Ile, Leu, Val, Phe.
26.2
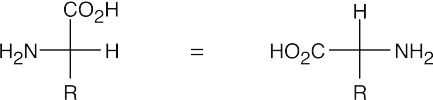
A Fischer projection of the α-carbon of an L-amino acid is pictured above.
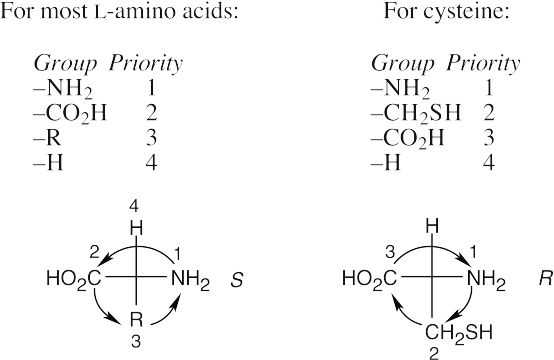
26.3
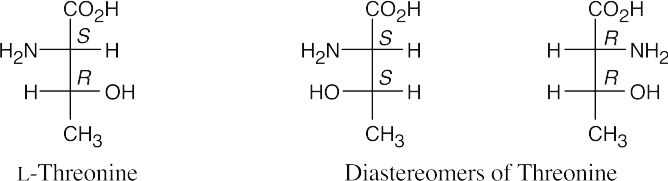
- On the low pH (acidic) side of pI, a protein has a net positive charge, and on the high pH (basic) side of pI, a protein has a net negative charge. Thus, hemoglobin (pI = 6.8) has a net positive charge at pH =5.3 and a net negative charge at pH = 7.3.
- This method of amino acid synthesis is simple and uses methods we have already studied. The phthalimide synthesis can also be used to introduce the amino group. Remember that only racemic amino acids are produced by this method.
 (a)
(a)
 (b)
(b)
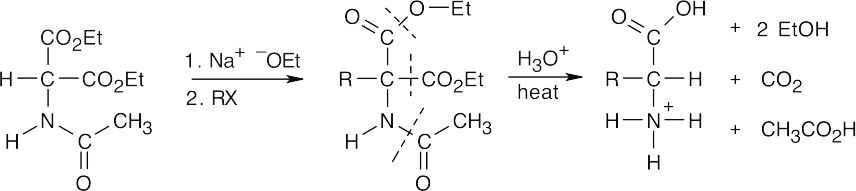
In the amidomalonate synthesis, shown above, an alkyl halide RX is converted to RCH(NH3+)CO2H. Choose an alkyl halide that completes the structure of the target amino acid.
 (a)
(a)
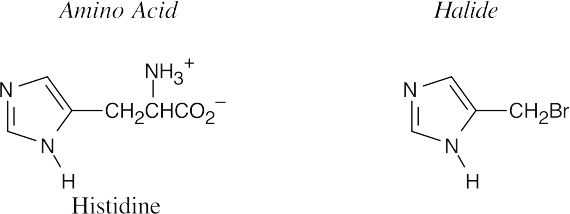 (b)
(b)
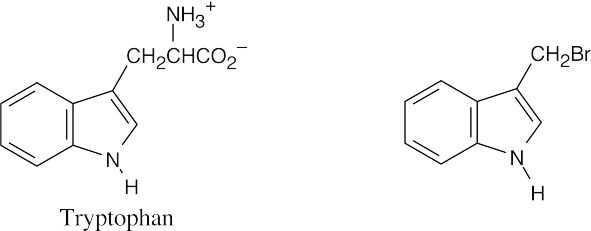 (c)
(c)
 (d)
(d)
- The precursor to an amino acid prepared by enantioselective hydrogenation has a Z
double bond conjugated with a carboxylic acid carbonyl group.
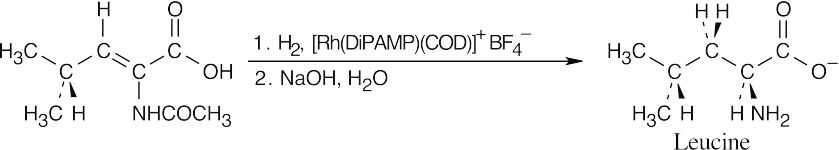
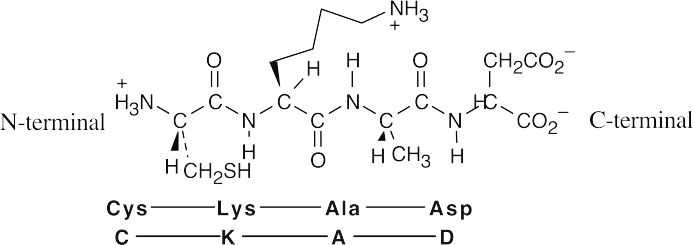
- Val–Tyr–Gly (VYG)Tyr–Gly–Val (YGV)Gly–Val–Tyr (GVY) Val–Gly–Tyr (VGY)Tyr–Val–Gly (YVG)Gly–Tyr–Val (GYV)
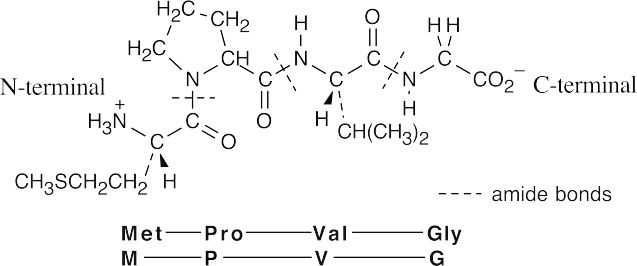

The cysteine sulfur is a good nucleophile, and iodide is a good leaving group.
- One product of the reaction of an amino acid with ninhydrin is the extensively conjugated purple ninhydrin product. The other major product is the aldehyde derived from the side chain of the amino acid. When valine reacts, the resulting aldehyde is 2-methylpropanal. The other products are carbon dioxide and water. The identity of the aldehyde is determined by the amino acid side chain.
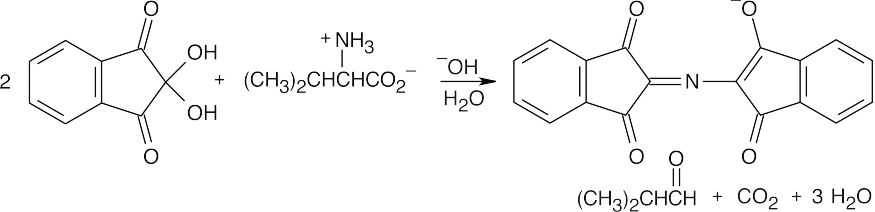
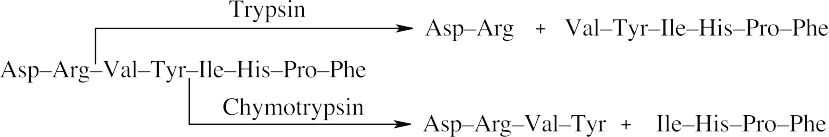
- Trypsin cleaves peptide bonds at the carboxyl (right) side of lysine and arginine. Chymotrypsin cleaves peptide bonds at the carboxyl side of phenylalanine, tyrosine and tryptophan.
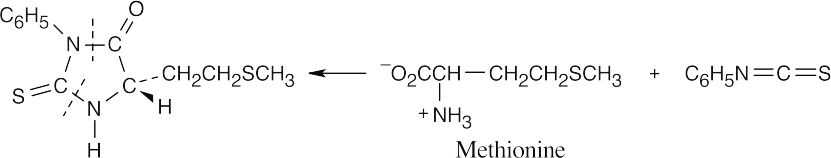
- The part of the PTH derivative that lies to the right of the indicated dotted lines comes from the N-terminal residue. Complete the structure to identify the amino acid, which in this problem is methionine.
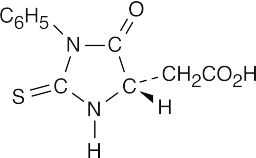
- The N-terminal residue of angiotensin II is aspartic acid. Replace the –R group of the PTH derivative in Figure 26.5 with –CH2CO2H to arrive at the correct structure.
- Line up the fragments so that the amino acids overlap.
|
(a)Arg–Pro |
(b) |
V–M–W |
|
Pro–Leu–Gly |
|
W–N-V |
|
Gly–Ile–Val The complete sequence: |
V–L |
|
|
|
The complete sequence: |
|
|
Arg–Pro–Leu–Gly–Ile–Val |
|
V–M–W-N–V–L |
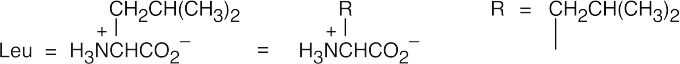
- Protect the amino group of leucine.
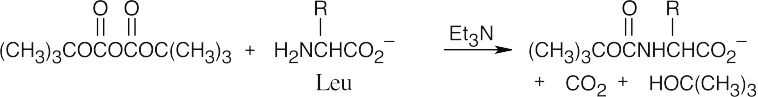
- Protect the carboxylic acid group of alanine.
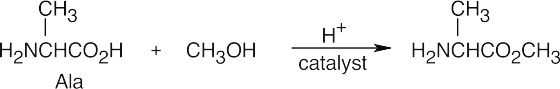
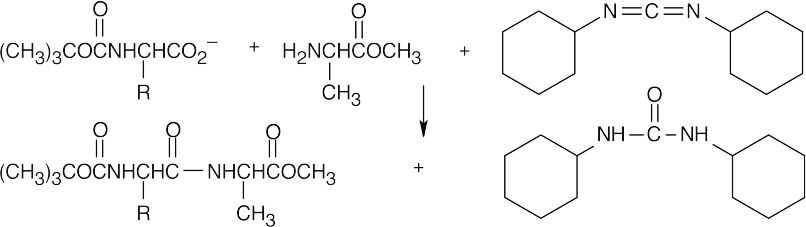
- Couple the protected amino acids with DCC.
- Remove the leucine protecting group.
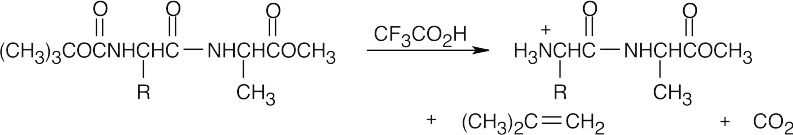
- Remove the alanine protecting group.
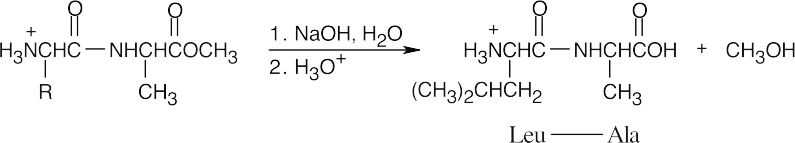
- Pyruvate decarboxylase is a lyase.
- Chymotrypsin is a hydrolase.
- Alcohol dehydrogenase is an oxidoreductase.
Additional Problems
Visualizing Chemistry
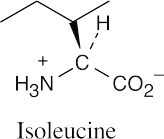
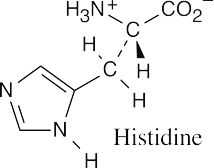
 (a)(b)(c)
(a)(b)(c)
- It’s possible to identify this representation of valine as the D enantiomer by noting the configuration at the chirality center. The configuration is R, and thus the structure is
D-valine.
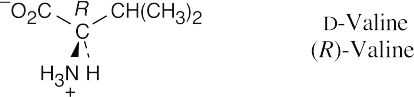
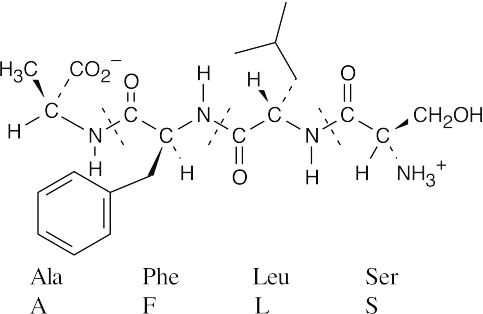
- After identifying the amino acid residues, notice that the tetrapeptide has been drawn with the amino terminal residue on the right. To name the sequence correctly, the amino terminal residue must be cited first. Thus, the tetrapeptide should be named Ser–Leu– Phe–Ala.
Mechanism Problems
26.24 (a)
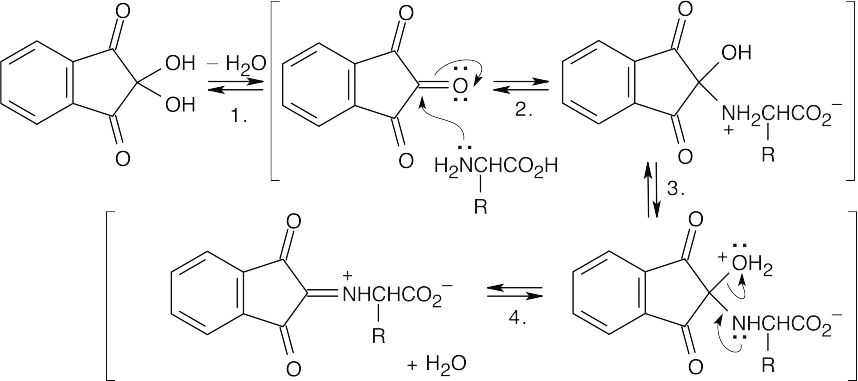
Step 1:Dehydration.
Step 2:Nucleophilic addition of the amino group of the amino acid.
Step 3:Proton transfer.
Step 4:Loss of water.
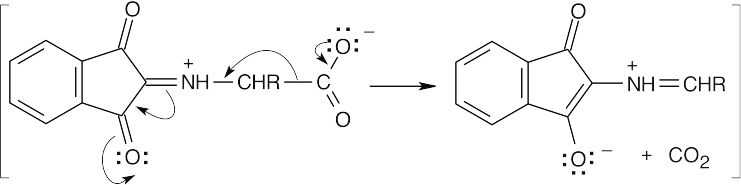 (b)
(b)
Decarboxylation produces a different imine.
(c)
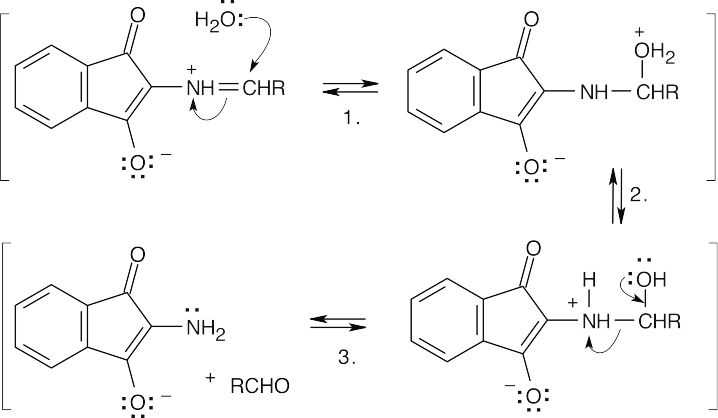
Step 1:Addition of water.
Step 2:Proton transfer.
Step 3:Bond cleavage to yield an aldehyde and an amine.
(d)
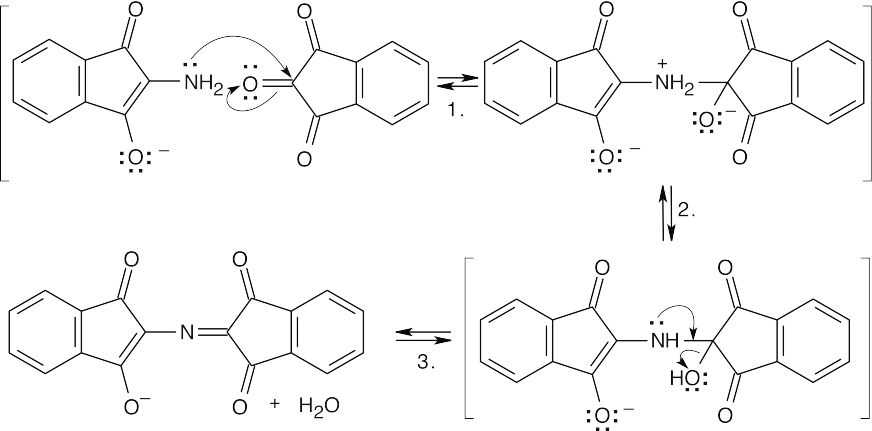
Step 1:Addition of the amine to a carbonyl carbon of a second ninhydrin molecule.
Step 2:Proton shift.
Step 3:Loss of water to form the purple anion.
Notice that the amino nitrogen is all that remains of the original amino acid.
26.25
Formation of cation:
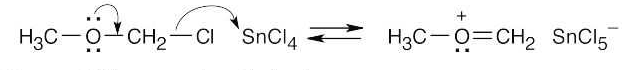
Electrophilic aromatic substitution:
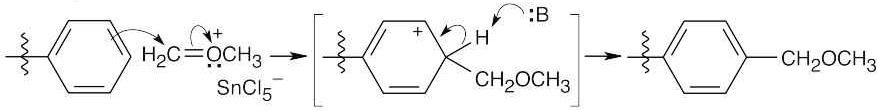
Protonation of the ether oxygen, followed by displacement of methanol by Cl–.
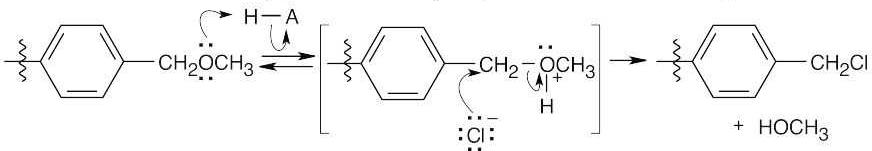
26.26
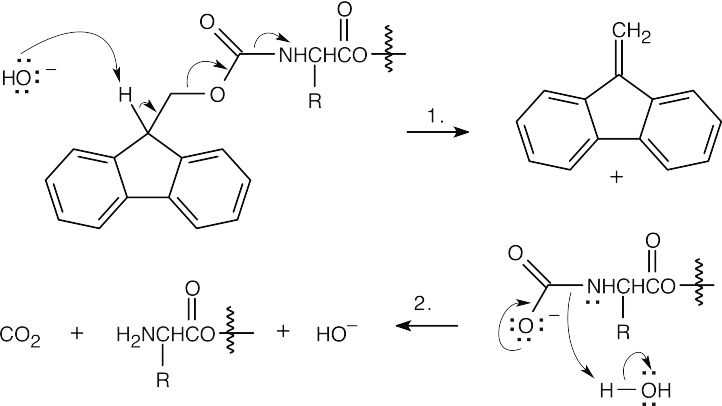
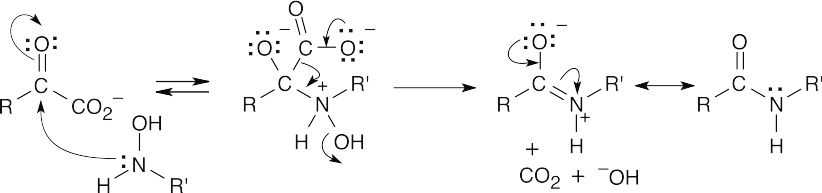
Step 1:NaOH brings about elimination of the carboxylated peptide.
Step 2:Loss of CO2.
The Fmoc group is acidic because the Fmoc anion is similar to the cyclopentadienyl anion, which is resonance-stabilized and is aromatic.
 26.27 (a)
26.27 (a)
The first step is a substitution similar to the nucleophilic acyl substitution reactions that we studied in Chapter 21.
(b)
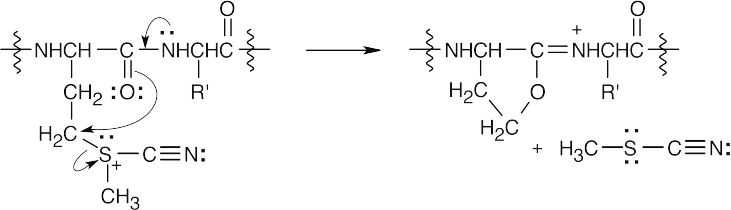
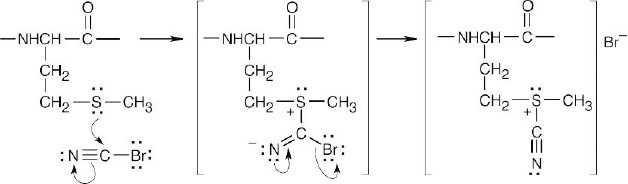
Internal SN2 displacement of sulfide results in formation of a 5-membered ring containing an iminium group.
(c)
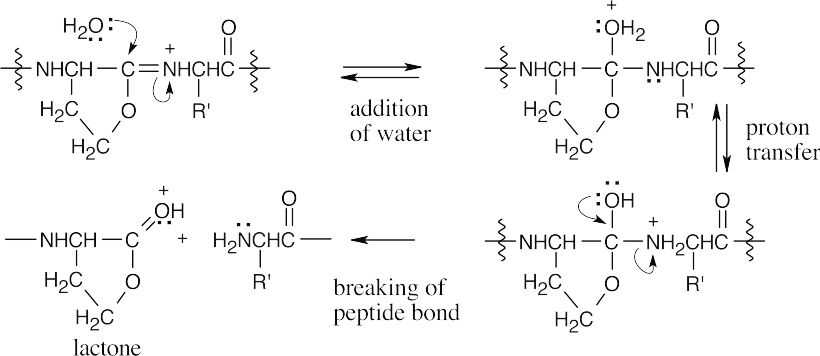
In this sequence of steps, water adds to the imine double bond, and the peptide bond is cleaved.
(d)
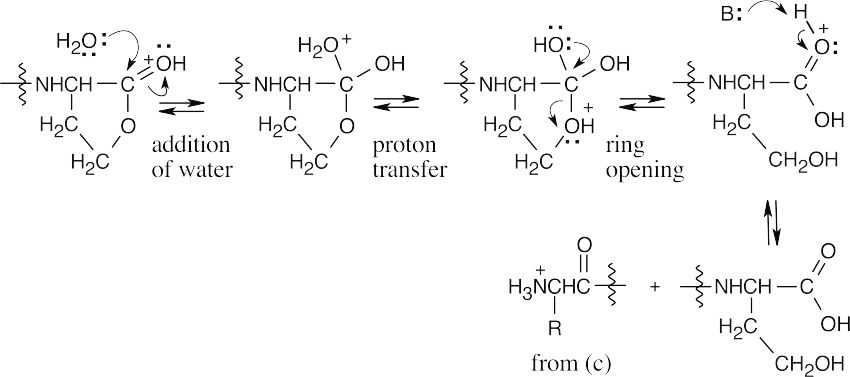
Water opens the lactone ring to give the product shown.
26.28
Amino Acid Structures and Chirality
26.29 Both (R)-serine and (R)-alanine are D-amino acids. In a D-amino acid, the –NH2 group is on the right.

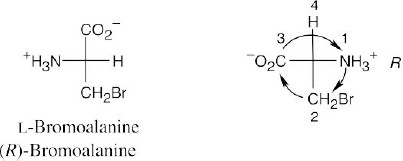 26.30
26.30
This L “amino acid” also has an R configuration because the –CH2Br “side chain” is
higher in priority than the –CO2H group.
26.31
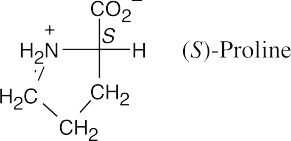
26.32
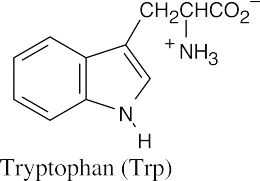
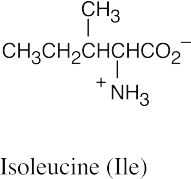
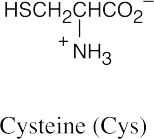 (b)(c)
(b)(c)
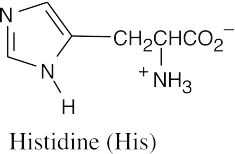 (d)
(d)
26.33
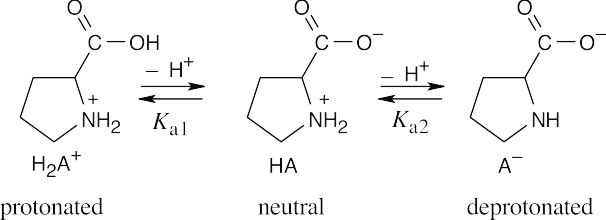
At pH = 2.50 :
log
[HA]
[H A+ ]
= pH − pKa1
= 2.50 −1.99 = 0.51;
[HA]
[H A+ ]
= 3.24
22
At pH = 2.50, approximately three times as many proline molecules exist in the neutral form as exist in the protonated form.
At pH = 9.70 :
[A− ][A− ]
log [HA] = pH − pKa2 = 9.70 −10.60 − 0.90; [HA] = 0.126
At pH = 9.70, the ratio of deprotonated proline to neutral proline is approximately 1:8.
|
26.34 (a)Val–Leu–Ser |
V-L-S |
Ser–Val–Leu |
S-V-L |
|
Val–Ser–Leu |
V-S-L |
Leu–Val–Ser |
L-V-S |
|
Ser–Leu–Val |
S-L-V |
Leu–Ser–Val |
L-S-V |
|
(b)Ser–Leu–Leu–Pro |
S-L-L-P |
Leu–Leu–Ser–Pro |
L-L-S-P |
|
Ser–Leu–Pro–Leu |
S-L-P-L |
Leu–Leu–Pro–Ser |
L-L-P-S |
|
Ser–Pro–Leu–Leu |
S-P-L-L |
Leu–Ser–Leu–Pro |
L-S-L-P |
|
Pro–Leu–Leu–Ser |
P-L-L-S |
Leu–Ser–Pro–Leu |
L-S-P-L |
|
Pro–Leu–Ser–Leu |
P-L-S-L |
Leu–Pro–Leu–Ser |
L-P-L-S |
|
Pro–Ser–Leu–Leu |
P-S-L-L |
Leu–Pro–Ser–Leu |
L-P-S-L |
- Aldehydes and ketones can undergo nucleophilic addition reactions. In particular, aldehydes and ketones can react with amines to form imines and enamines, reactions that might compete with formation of amide bonds between amino acids. Because of this reactivity, aldehydes and ketones are unlikely to be found in amino acid sidechains.
Amino Acid Synthesis and Reactions
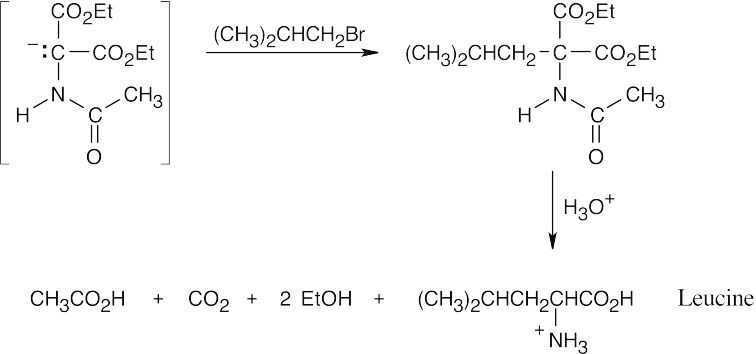
- The diethylamidomalonate anion is formed by treating diethylamidomalonate with sodium ethoxide. Choose the appropriate halide based on the amino acid side chain.
(a)
(b)
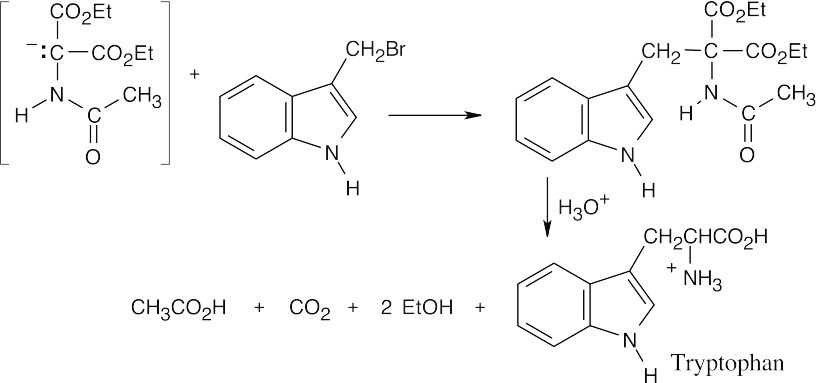
26.37
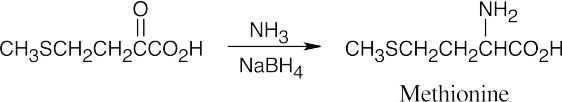 (a)
(a)
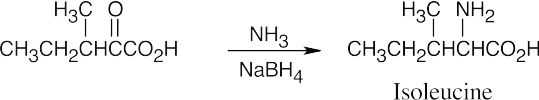 (b)
(b)
26.38
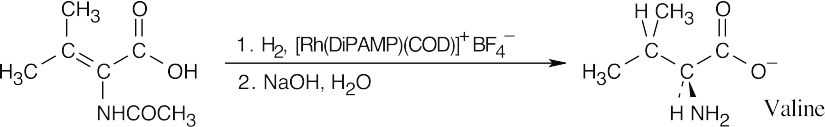 (a)
(a)
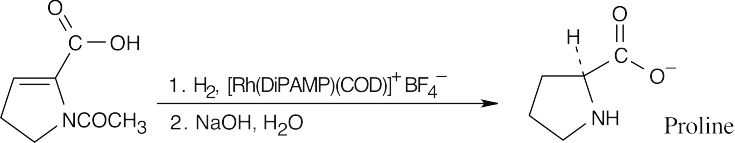 (b)
(b)
26.39
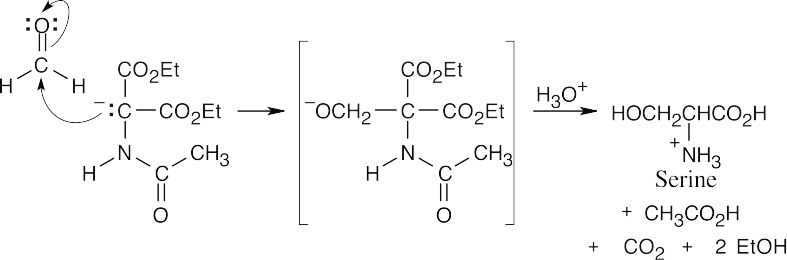
26.40
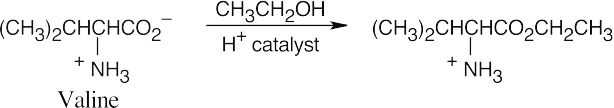 (a)
(a)
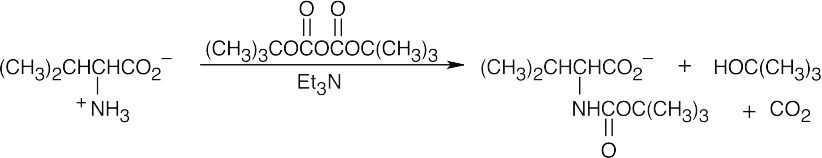 (b)
(b)
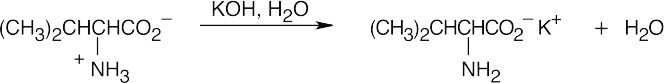 (c)
(c)
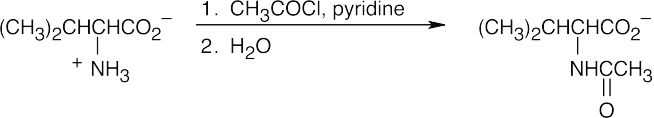 (d)
(d)
26.41
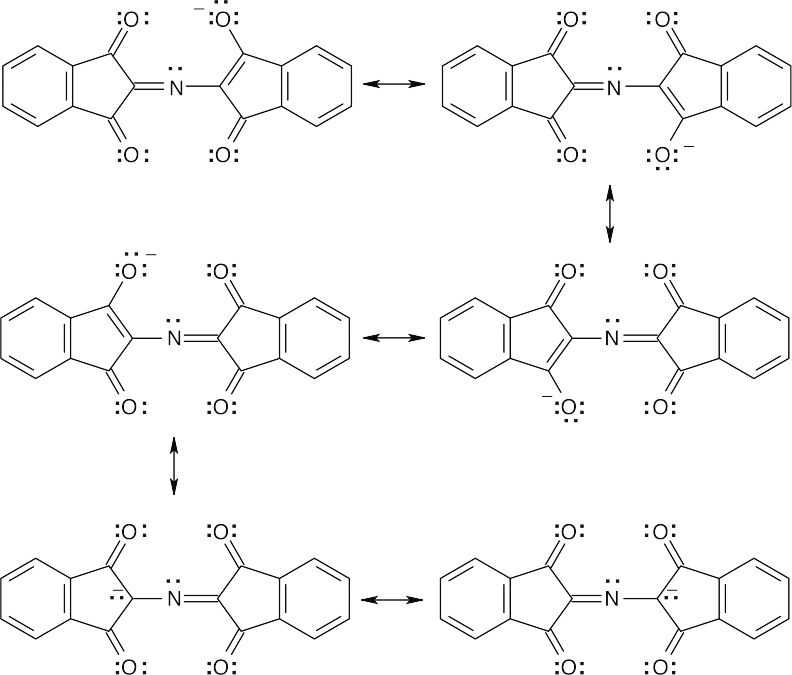
It is also possible to draw many other resonance forms that involve the π electrons of the aromatic 6-membered rings.
Peptides and Enzymes
26.42 (a)
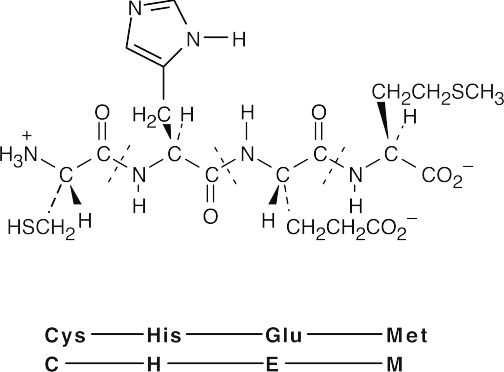
(b)
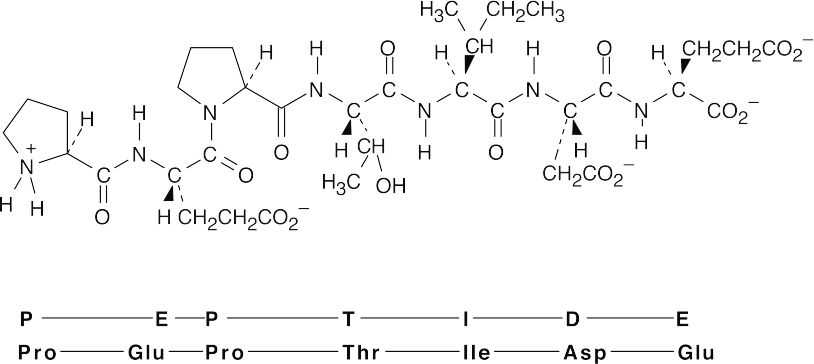
26.43 The tripeptide is cyclic.

26.44
Step 1: Valine is protected as its Boc derivative.
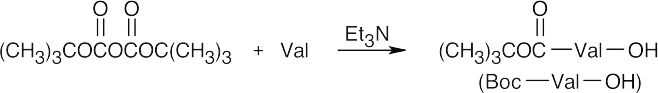
Step 2: Boc–Val bonds to the polymer in an SN2 reaction.
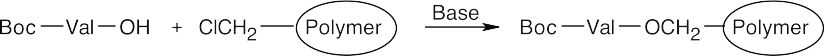
Step 3: The polymer is first washed, then is treated with CF3CO2H to cleave the Boc group.

Step 4: A Boc-protected Ala is coupled to the polymer-bound valine by reaction with DCC. The polymer is washed.

Step 5: The polymer is treated with CF3CO2H to remove Boc.

Step 6: A Boc-protected Phe is coupled to the polymer by reaction with DCC. The polymer is washed.
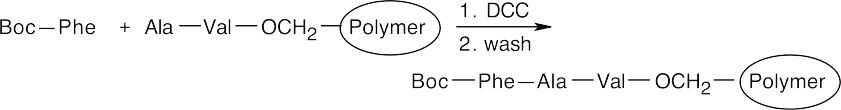
Step 7: Treatment with anhydrous HF removes the Boc group and cleaves the ester bond between the peptide and the polymer.
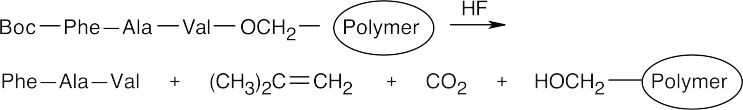
26.45
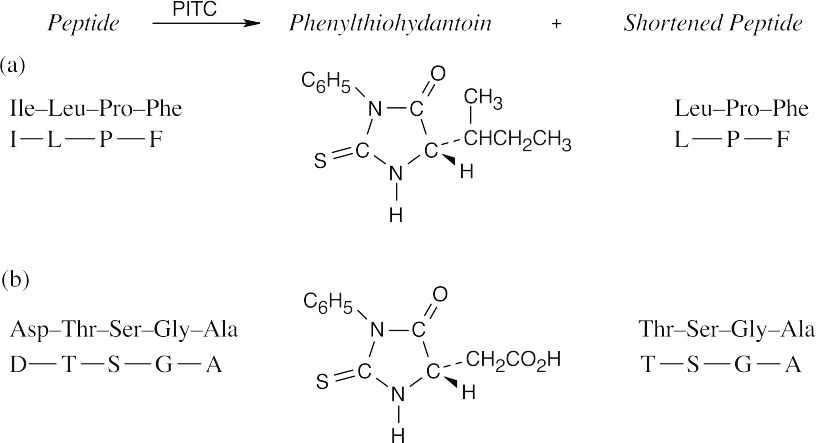
26.46
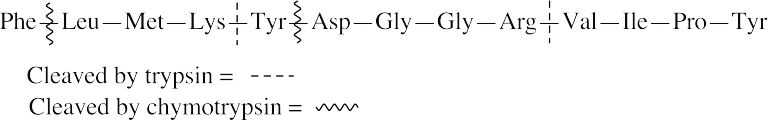
- (a)Hydrolases catalyze the cleavage of bonds by addition of water (hydrolysis).
- Lyases catalyze the elimination of a small molecule (H2O, CO2) from amolecule.
- Transferases catalyze the transfer of a functional group between substrates.
- Amino acids with polar side chains are likely to be found on the outside of a globular protein, where they can form hydrogen bonds with water and with each other. Amino acids with nonpolar side chains are found on the inside of a globular protein, where they can avoid water. Thus, aspartic acid (b) and lysine (d) are found on the outside of a globular protein, and valine (a) and phenylalanine (c) are likely to be found on the inside. Refer to Table 26.1.
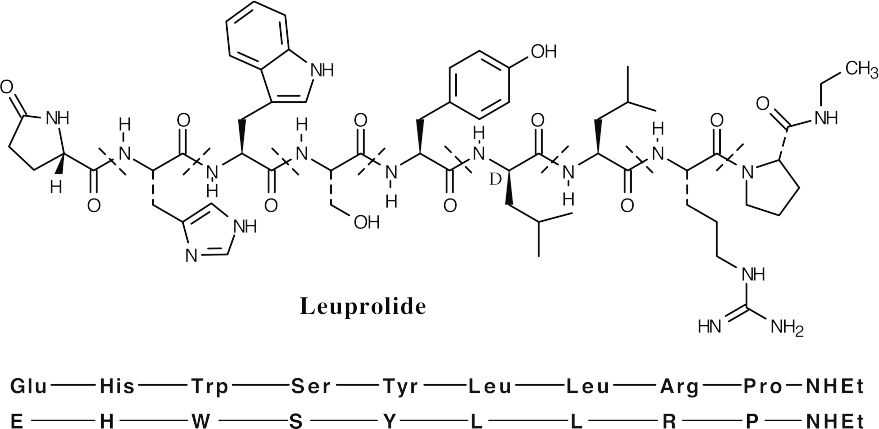
- The N-terminal glutamic acid is a cyclic lactam. The C-terminal proline is an N– ethyl amide.
- One of the leucines (indicated above) has D stereochemistry.
- See above.
- The charge on a peptide is due to the side chains. According to Table 26.1, the only side chain that is charged at neutral pH is arginine. Thus, leuprolide has a charge of
+1 at neutral pH.
General Problems
- A proline residue in a polypeptide chain interrupts α-helix formation because the amide nitrogen of proline has no hydrogen that can contribute to the hydrogen-bonded structure of an α-helix.
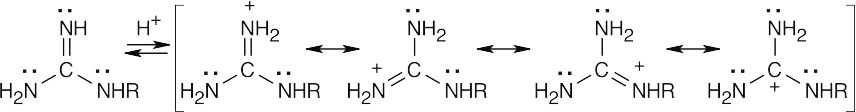
The protonated guanidino group can be stabilized by resonance.
- 100 g of cytochrome c contains 0.43 g iron, or 0.0077 mol Fe:
0.43 g Fe× 1 mol Fe
55.8 g Fe
= 0.0077 mol Fe
Assuming that each mole of protein contains 1 mol Fe, then mol Fe = mol protein.
100 g Cytochrome c = 13,000 g Cytochrome c
0.0077 mol Fe1 mol Fe
Cytochrome c has a minimum molecular weight of 13,000 g/ mol.
- 1H NMR shows that the two methyl groups of N,N-dimethylformamide are nonequivalent at room temperature. If rotation around the CO–N bond were unrestricted, the methyl groups would be interconvertible, and their 1H NMR absorptions would coalesce into a single signal.
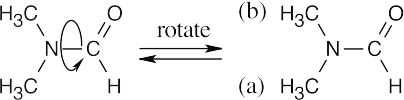
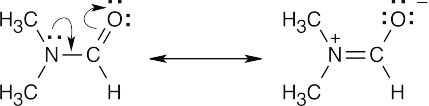
The presence of two methyl absorptions shows that there is a barrier to rotation around the CO–N bond. This barrier is due to the partial double-bond character of the CO–N bond, as indicated by the two resonance forms below. Rotation to interconvert the two methyl groups is slow at room temperature, but heating to 180° supplies enough energy to allow rapid rotation and to cause the two NMR absorptions to merge.
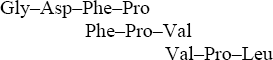
The complete sequence: Gly–Gly–Asp–Phe–Pro–Val–Pro–Leu
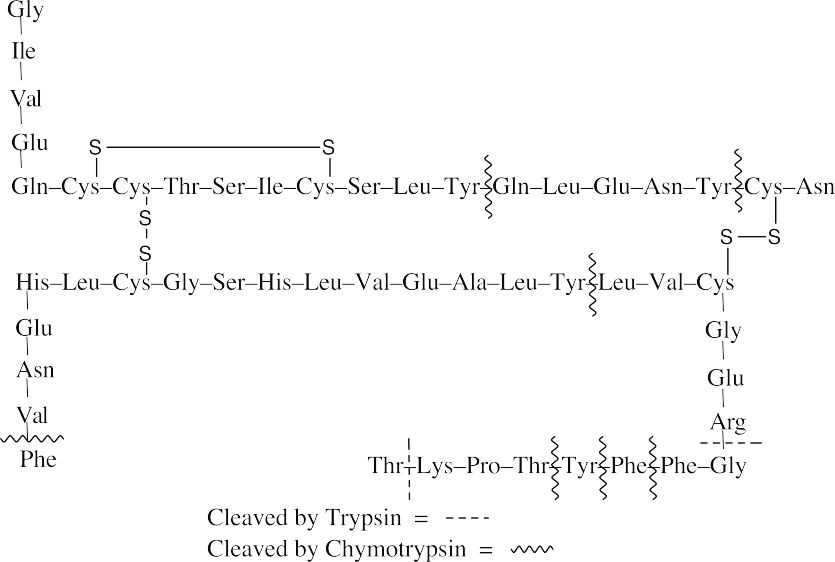
- Ser–Ile–Arg–Val–Val–Pro–Tyr–Leu–Arg
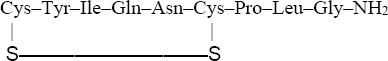 Reduced oxytocin:Cys–Tyr–Ile–Gln–Asn–Cys–Pro–Leu–Gly–NH2 Oxidized oxytocin:
Reduced oxytocin:Cys–Tyr–Ile–Gln–Asn–Cys–Pro–Leu–Gly–NH2 Oxidized oxytocin:
The C-terminal end of oxytocin is actually an amide, but this can’t be determined from
the information given.
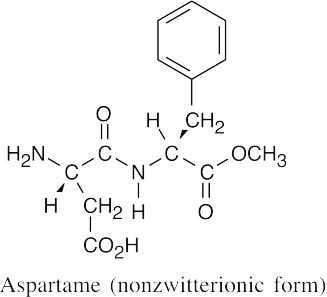
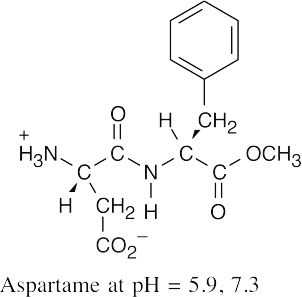 (b)
(b)
At pH = 7.3, aspartame exists in the zwitterionic form, as it does at pH = 5.9.
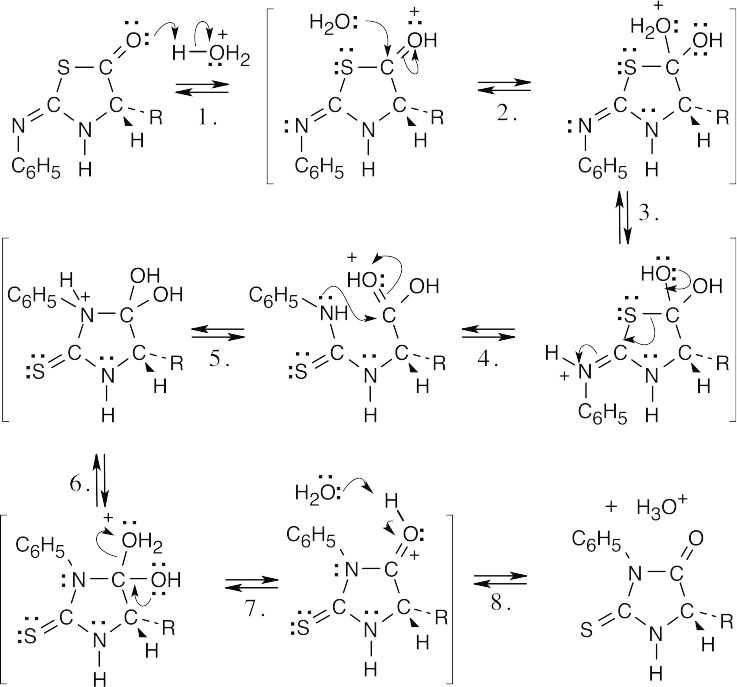
|
Step 1: |
Protonation. |
Step 2: |
Addition of water. |
|
Step 3: |
Proton transfer. |
Step 4: |
Ring opening. |
|
Step 5: |
Bond rotation, addition of amine. |
Step 6: |
Proton transfer. |
|
Step 7: |
Loss of water. |
Step 8: |
Deprotonation. |

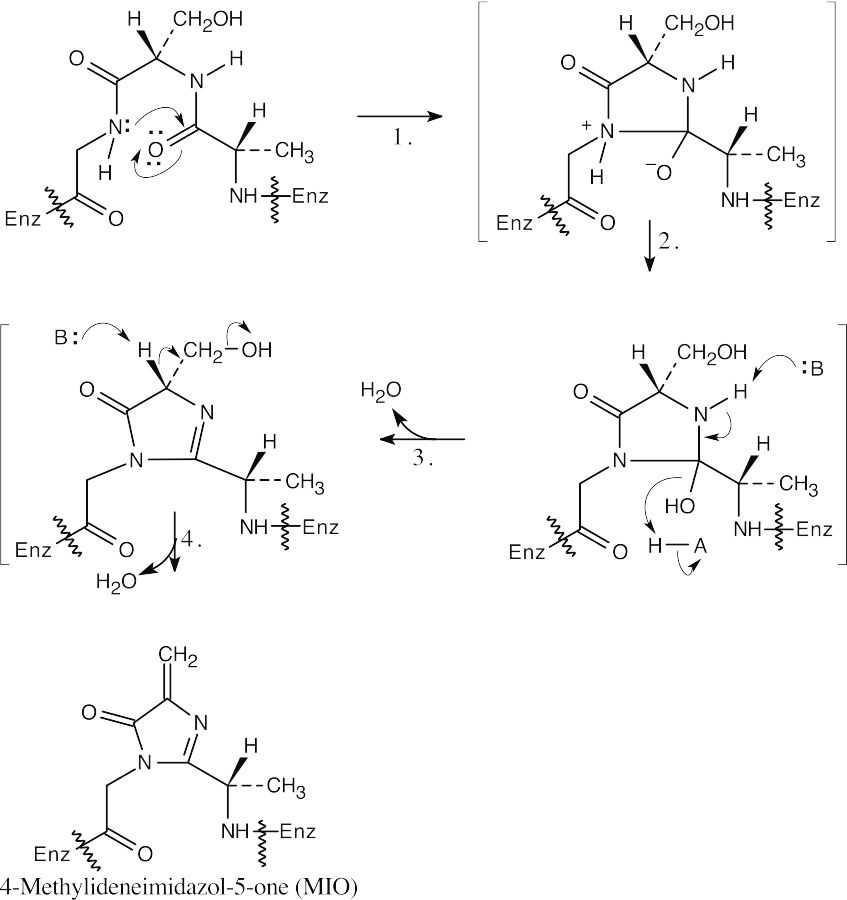
Step 1:Nucleophilic addition of the amino group of the amino acid.
Step 2:Proton transfer.
Step 3:Elimination.
Step 4:Elimination.
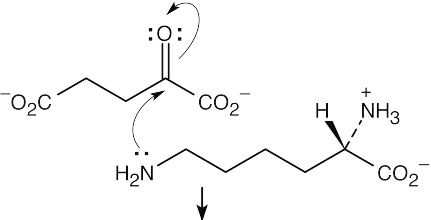
Step 1:Nucleophilic addition of the amine to α-ketoglutarate.
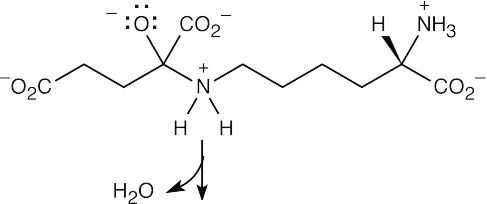
Step 2:Loss of water.
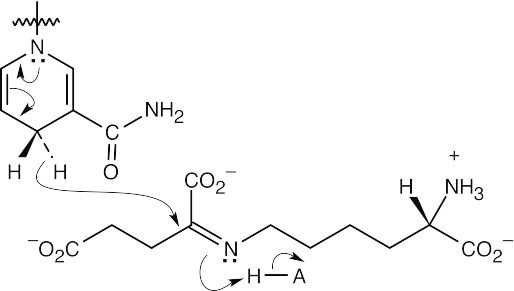
Step 3:Reduction by NADPH/H+
This file is copyright 2023, Rice University. All Rights Reserved.