6 Genome Evolution
By the end of this chapter you will be able to:
- Describe the ways in which organisms have different genomes
- Explain how new genes arise
- Describe how plants grow vegetatively
- Explain how transposons influence gene expression
- Describe exon shuffling
- Compare and contrast a gene and a pseudogene
Introduction
Genome evolution refers to the process of changes in the genetic material (DNA) of an organism over time. The totality of DNA found in a eukaryotic organism, all the DNA in the nuclei of cells, in the mitochondria, and if a plant in the chloroplasts, is referred to as the genome. The DNA is bound with proteins to form complexes called chromosomes. Each species of eukaryote has a characteristic number of chromosomes in the nuclei of its cells. Human body cells (somatic cells) have 46 chromosomes. A somatic cell contains two matched sets of chromosomes, a configuration known as diploid (symbol is 2n). Human cells that contain one set of 23 chromosomes are called gametes, or sex cells; these eggs and sperm are designated n, or haploid.
The genome of bacteria and prokaryotes includes the DNA sequence in the cell as well as any DNA sequences of plasmids, which are extra pieces of DNA that are not essential for normal growth. In prokaryotes, the region in the cell that contains the DNA molecule in the form of a loop or circle is called the nucleoid.
The genome of organisms consists of DNA that contains both exons, which code for proteins, and introns, referred to as repetitive DNA. All species have a characteristic genome size ranging from several thousand in bacteria to 20 – 30 thousand in more complex organisms (Fig 1).
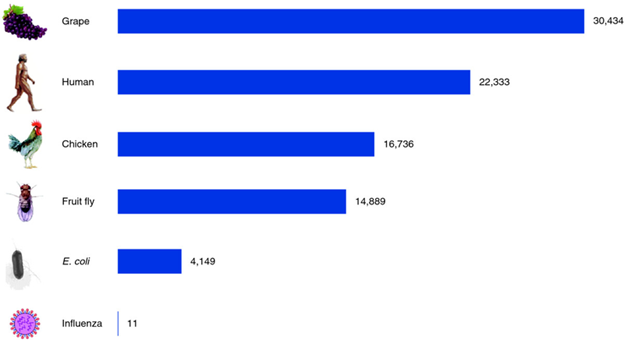
Figure 1: Gene counts in a variety of species. Viruses, the simplest particles, have only a handful of genes. Bacteria such as Escherichia coli have a few thousand genes, and multicellular plants and animals have two to ten times more. The chicken and grape gene counts shown here are based on draft genomes [50, 51] and may be revised substantially in the future. Pertea M, Salzberg SL. Between a chicken and a grape: estimating the number of human genes. Genome Biology. 2010;11(5):206. doi:10.1186/gb-2010-11-5-206. https://genomebiology.biomedcentral.com/articles/10.1186/gb-2010-11-5-206 From https://commons.wikimedia.org/wiki/File:Gene_counts_in_a_variety_of_species.png
Several key mechanisms contribute to genome evolution, including mutation, natural selection, genetic drift, horizontal gene transfer, and gene duplication. Mutation, natural selection, and genetic drift have been discussed in previous chapters. This chapter will discuss additional concepts, including gene transfer and gene duplication.
Sources of New Genes
Within One Organism
There are surprisingly more ways to create a new gene besides DNA mutation, although a change in a DNA sequence that leads to a change in the protein is one way to find a new gene or trait (Fig 2).
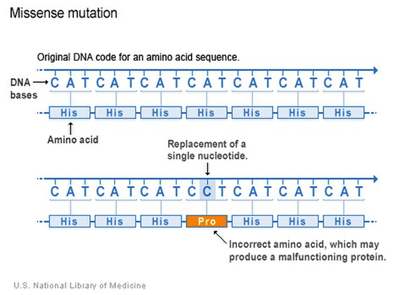
Figure 2: A section of DNA showing the replacement of a single nucleotide from adenine to cytosine, a point mutation, leading to a different and incorrect amino acid (shown in orange) being incorporated into the protein sequence. U.S. National Library of Medicine, Public domain, via Wikimedia Commons. https://commons.wikimedia.org/wiki/File:Missense_Mutation_Example.jpg
Recall that non-coding or repetitive sections of DNA called introns can mutate without consequence and can do so at a predictable rate (i.e. molecular clock). These non-coding regions do not code for a particular protein and therefore any change in them is unusually not impactful. Again, this is inconsequential because the introns do not code for a specific protein. Changes to the intron sequence should not have an effect, until they do influence the expression of the exons or if enough changes occur that the intron is no longer an intron and acts as an exon. The scientific community has discovered at least 155 human genes that have evolved from introns. Because these new genes are so small in length (about 300 nucleotides long) they are called microgenes which likely explains why they were overlooked for so long (Vakirlis et al 2022).
Not only can genes evolve, but so too can chromosomes. Whole chromosomes or segments of chromosomes may be duplicated, or segments flipped around and inverted, thereby changing messages and genes. Such interchromosomal rearrangements can lead to speciation events in a variety of organisms, such as fish. Research has shown that eight major interchromosomal rearrangements occurred soon after fish-specific whole genome duplication in an ancestor that led to species of pufferfish, zebrafish, and medaka fish (Kasahara et al 2007).
Genes can also get flipped or rearranged on a chromosome or even jump from one section of a chromosome to another section or a different chromosome altogether. These genes are commonly referred to as jumping genes for their behavior or transposons. You may also hear these jumping genes called transposable elements. In some cases, the transposons affect gene expression but they can also create a new gene if the transposon stays in place and serves a new function.
The first transposons were discovered in the 1940s by Barbara McClintock who worked with maize (Zea mays corn). She found that they were responsible for a variety of types of gene mutations. In developing somatic tissues like corn kernels, a mutation (e.g., “c” mutation) that alters color will be passed on to all the descendant cells. This produces the variegated pattern which is so prized in “Indian corn” (Fig 3). It took about 40 years for other scientists to fully appreciate the significance of Barbara McClintock’s discoveries. She was finally awarded a Nobel Prize in 1983.

Figure 3: Indian corn. The color of the kernels is the result of transposons or jumping genes. Sam Fentress, CC BY-SA 2.0 <https://creativecommons.org/licenses/by-sa/2.0>, via Wikimedia Commons. https://commons.wikimedia.org/wiki/File:Corncobs.jpg
Transposons are activated by stress, such as changes in temperature, oxygen, toxins, or nutrients, which can result in new genetic variations. This new variation can lead to the evolution of traits. For example, transposons account for at least half of all phenotype-altering mutations in flies where infected with the pathogen stressor of Pseudomonas. Once the transposable elements moved, variant flies were produced that could survive in this environment (Rech et al 2019). In another example, Anopheles coluzzii mosquitoes in new habitats with new stressors, have been found to show transposable elements inserted near genes involved in insecticide resistance and immunity, helping to drive the mosquitoes’ ecological flexibility and their ability to live in these new environments (Vargas-Chavez et al. 2021).
Genes can not only move from one location to another, parts of them, that is their exons, can also get shuffled within their original location to produce new combinations of messages and therefore new traits or proteins. This exon recombination has led to the production of a variety of antibodies, proteins that can attach to foreign pathogens such as viruses in human cells and thereby render them ineffective (until the rest of the immune system can destroy the pathogens).
Susumu Tonegawa was awarded the Nobel Prize in Physiology or Medicine for this discovery of the genetic principle for the generation of antibody diversity. In 1983 he published his research showing how bone marrow-derived lymphocytes (white blood cells) use segments of DNA that have been recombined in ways to make a new complete gene, which contributes to increasing the diversity of antibodies that can be produced by a single organism (Tonegawa 1983).
Between Organisms
Horizontal gene transfer is the process by which genetic material (DNA or sometimes RNA) is transferred between different, often unrelated, organisms. Unlike the more common vertical gene transfer, which occurs from parent to offspring during reproduction, horizontal gene transfer allows for the exchange of genetic material between individuals that may not be directly related by descent.
Horizontal gene transfer can happen between organisms of the same species (intraspecific) or between different species (interspecific). It is most prevalent in prokaryotes, such as bacteria and archaea, but has also been observed in some eukaryotic organisms. It is quite common for virus DNA to be interspecifically transferred to another species.
There are several mechanisms through which horizontal gene transfer can occur between species:
- Transformation: In transformation, bacteria or other organisms take up free DNA from their surroundings. This DNA can come from dead or lysed cells, and if it contains functional genes, it may be integrated into the recipient organism’s genome.
- Transduction: Transduction is a process in which genetic material is transferred from one bacterium to another through a bacteriophage (a type of virus that infects bacteria). During the viral replication cycle, the phage may accidentally package a fragment of the host’s DNA and transfer it to another bacterium when it infects it.
Horizontal gene transfer has significant implications for the evolution and diversification of organisms. It allows for the rapid acquisition of new traits and genes, enabling species to adapt to changing environments or new ecological niches more efficiently. In prokaryotes, horizontal gene transfer has played a crucial role in the spread of antibiotic resistance genes, making it a critical concern in the context of antibiotic resistance in pathogens.
In eukaryotes, horizontal gene transfer is relatively less common but still occurs, particularly in simpler organisms like fungi, plants, and protists. While it might not be as prevalent or significant as in prokaryotes, the transfer of genes between eukaryotic species can still influence their evolutionary trajectories and contribute to the genetic diversity observed in these organisms.
Duplication of Genes & Pseudogenes
Gene duplication occurs when a segment of DNA is copied, resulting in multiple copies of a gene within a genome. These duplicated genes can diverge over time, leading to the evolution of new functions or specialization, contributing to the complexity and diversity of organisms.
We can also see what looks to be a new gene, say a copy or duplication of an existing gene, but then it doesn’t do anything. When you look closer at this “new” gene, you can see what appears to be exons and introns (sections of repetitive DNA); however, it turns out that this gene doesn’t work. This would be a pseudogene.
Because pseudogenes do not affect function, mutations in pseudogenes tend to be neutral and they can accumulate rapidly over evolutionary time (the molecular clock).
The scientific community continues to learn more about pseudogenes and we are learning that in some cases, pseudogenes are not as inactive as we thought. It has been shown that pseudogenes are capable of dampening cancer-causing genes, called oncogenes, and therefore suppressing tumors (Pink et al 2011).
Genome Increases in Plants
As new genes emerge and as changes occur to the genome, we can see speciation events arising as well. Plants in particular seem to be able to handle large increases in their genome size compared to animals. We have seen in evolutionary history some plants have three or more sets of chromosomes, rather than the normal two copies of chromosomes. That is they are polyploidy, not diploid. We have evidence that whole genome duplications have occurred in plants that cluster around a mass extinction event around 65 million years ago. Whole genome duplication has been linked to major evolutionary transitions such as the diversification of flowering plants (e.g. Soltis et al 2014) with approximately 35% of all flowering species as recent polyploid and 15% of speciation events in angiosperms directly associated with genome duplication (Wood et al 2009).
Although scientists still have a lot to learn about why and how plants can handle such large loads of DNA, there is a thought that their likely success lies in their vegetative capabilities. Vegetative growth is the period of growth between germination and flowering. This is when a plant will rapidly produce more genetically identical cells through mitosis to grow taller or wider. Plants, therefore, can rapidly grow (longer stems or roots) given the right environmental conditions.
During vegetative growth, a plant uses its meristematic tissue (permanently embryonic tissue that can constantly undergo cell division) for rapid mitotic division. Numerous plant physiological processes such as photosynthesis, cell stretching, transport of sugars, and mitosis are called into play. Meristems are cells that are permanently embryonic and found at the tips of shoots and roots. These embryonic cells are “primed” to engage in mitosis over and over again quickly, which leads to rapid growth for plants. Meristems are analogous to stem cells in animals – these are cells that can quickly divide to give rise to new cells. For example, you might have an apical stem meristem at the bud of a plant giving rise to new leaves.
For a great time-lapse motion sequence of a tulip tree and its leaf bud growth, click here: Plants In Motion (indiana.edu)
Most plants will grow vegetatively and continually throughout their lives. They grow through a combination of cell growth and cell division (mitosis). Meristems allow plant roots and stems to grow longer in a process called primary growth and they can also grow wider in a process called secondary growth, which is often referred to as wood. Often the rate of growth and the timing of growth are connected to environmental stimuli and environmental conditions. A plant’s sensory response to external stimuli relies on chemical messengers (hormones). Plant hormones affect all aspects of plant life, from flowering to fruit setting and maturation, and from phototropism to leaf fall. Potentially every cell in a plant can produce plant hormones. They can act in their cell of origin or be transported to other portions of the plant body, with many plant responses involving the synergistic or antagonistic interaction of two or more hormones. In contrast, animal hormones are produced in specific glands and transported to a distant site for action, and they act alone.
Plants can also self-fertilize. This means that once a change in ploidy has occurred, an individual doesn’t need to rely on finding another mate with the same number of chromosomes as itself. This can lead to a high frequency of polyploid speciation in many plant lineages. For example, approximately 31% of all fern species have originated as a direct result of polyploid speciation (Wood et al 2009).
In general, for all plants, when the environmental conditions are stable with shade, plants seem to be able to handle increased loads of DNA and increase their genome size. Plants growing in unstable and sunny environments do not seem as successful with incorporating polyploidy. This may or may not be related to the type of plant. Here, we can also see a connection with generation time in plants. The generation time hypothesis that the rate of evolution is faster for shorter-lived species of plants compared to longer-lived species is supported by the data. Herbaceous groups of plants have median rates of evolution 2.7 to 10 times as high as the median rates in shrub and tree species (Smith & Donoghue 2008).
Summary
Genome evolution refers to the process of changes in the genetic material (DNA) of an organism over time and is a fundamental aspect of biological evolution, playing a crucial role in shaping the diversity of life on Earth. Several key mechanisms contribute to genome evolution, including jumping genes, horizontal gene transfer, and gene duplication.
Genome evolution plays a crucial role in the adaptation of organisms to changing environments and ecological niches. It drives speciation, the process by which new species arise, and is responsible for the vast array of life forms found on our planet. The study of genome evolution helps us understand the genetic basis of traits, diseases, and the relationships between different species, providing valuable insights into the history and mechanisms of life’s development. Advances in genomics and bioinformatics have significantly improved our understanding of genome evolution and continue to unravel the complexities of this fascinating field.
Questions
Glossary
References
Kasahara, M., Naruse, K., Sasaki, S. et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447, 714–719. https://doi.org/10.1038/nature05846
Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, & DRF Carter. 2011. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 17(5): 792-798.
Rech GE, Bogaerts-Marquez M, Barron MG, Merenciano M, Villanueva-Canas JL, Horvath V, Fiston-Lavier AS,, Luyten I, Venkataram S, Quesneville H, & DA Petrov. 2019. Stress response, behavior and development are shaped by transposable element-induced mutations in Drosophila. PLoS Genetics 15(2): e1007900 https://doi.org/10.1371/journal.pgen.1007900
Smith, S & M Donoghue. 2008. Rates of molecular evolution are linked to life history in flowering plants. Science 322: 86-89.
Soltis, D. E., P. S. Soltis, J. C. Pires, A. Kovarik, J. A. Tate, and E. Mavrodiev. 2004. Recent and recurrent polyploidy in Tragopogon (Asteraceae): cytogenetic, genomic and genetic comparisons. Biol. J. Linn. Soc. 82: 485– 501.
Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature 302: 575-581.
Vakirlis N, Vane Z, Duggan KM, & A McLysaght. 2022. De novo birth of functional microproteins in the human lineage. Cell Reports 41 (12): 111808. https://doi.org/10.1016/j.celrep.2022.111808.
Vargas-Chavez C, Long Pendy NM, Nsango SE, Aguilera L, Ayala D, & J. Gonzalez. 2021. Transposable element variants and their potential adaptive impact in urban populations of the malaria vector Anopheles coluzzii. Genome Research 32: 189-202. doi:10.1101/gr.275761.121
Wood, T. E., N. Takebayashi, M. S. Barker, I. Mayrose, P. B. Greenspoon, and L. H. Rieseberg. 2009. The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 106: 13875– 13879.

