28.8 The Polymerase Chain Reaction
It often happens that only a tiny amount of DNA can be obtained directly, as might occur at a crime scene, so methods for obtaining larger amounts are sometimes needed to carry out sequencing and characterization. The invention of the polymerase chain reaction (PCR) by Kary Mullis in 1986 has been described as being to genes what Gutenberg’s invention of the printing press was to the written word. Just as the printing press produces multiple copies of a book, PCR produces multiple copies of a given DNA sequence. Starting from less than 1 picogram (pg) of DNA with a chain length of 10,000 nucleotides (1 pg = 10–12 g; about 105 molecules), PCR makes it possible to obtain several micrograms (1 μg = 10–6 g; about 1011 molecules) in just a few hours.
The key to the polymerase chain reaction is Taq DNA polymerase, a heat-stable enzyme isolated from the thermophilic bacterium Thermus aquaticus found in a hot spring in Yellowstone National Park. Taq polymerase is able to take a single strand of DNA having a short, primer segment of complementary chain at one end and then finish constructing the entire complementary strand. The overall process takes three steps, as shown in Figure
28.10. More recently, improved heat-stable DNA polymerases have become available, including Vent polymerase and Pfu polymerase, both isolated from bacteria growing near geothermal vents in the ocean floor. The error rate of both enzymes is substantially less than that of Taq.
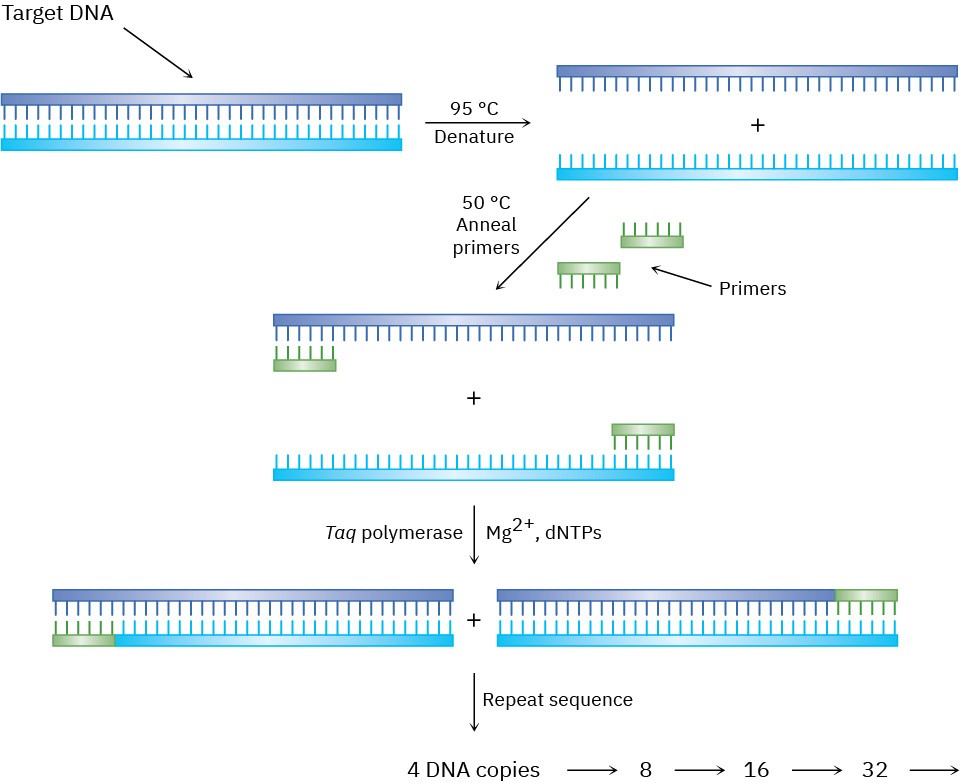
Figure 28.10 The polymerase chain reaction. Details are explained in the text.
STEP 1
The double-stranded DNA to be amplified is heated in the presence of Taq polymerase, Mg2+ ion, the four deoxynucleotide triphosphate monomers (dNTPs), and a large excess of two short oligonucleotide primers of about 20 bases each. Each primer is complementary to the sequence at the end of one of the target DNA segments. At a temperature of 95 °C, double-stranded DNA denatures, spontaneously breaking apart into two single strands.
STEP 2
The temperature is lowered to between 37 and 50 °C, allowing the primers, because of their relatively high concentration, to anneal by hydrogen-bonding to their complementary sequence at the end of each target strand.
STEP 3
The temperature is then raised to 72 °C, and Taq polymerase catalyzes the addition of further nucleotides to the two primed DNA strands. When replication of each strand is complete, two copies of the original DNA now exist. Repeating the denature–anneal– synthesize cycle a second time yields four DNA copies, repeating a third time yields eight copies, and so on, in an exponential series.
PCR has been automated, and 30 or so cycles can be carried out in an hour, resulting in a theoretical amplification factor of 230 (∼109). In practice, however, the efficiency of each cycle is less than 100%, and an experimental amplification of about 106 to 108 is routinely achieved for 30 cycles.
Additional Problems 28 • Additional Problems 28 • Additional Problems Visualizing Chemistry Problem 28-13
Identify the following bases, and tell whether each is found in DNA, RNA, or both:
(a)
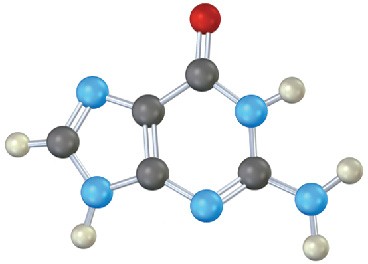
(b)
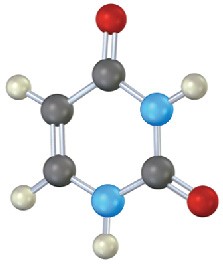
(c)
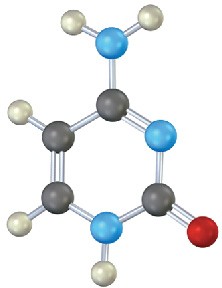
Problem 28-14
Identify the following nucleotide, and tell how it is used:
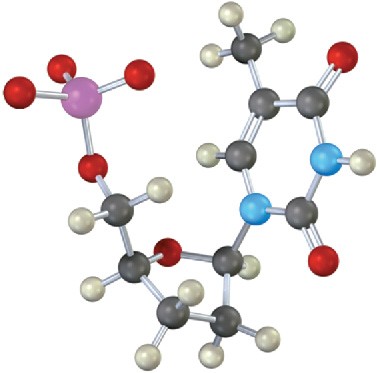
Problem 28-15
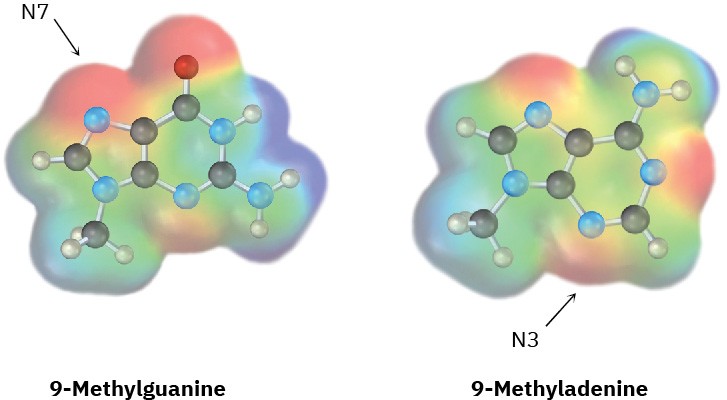
Amine bases in nucleic acids can react with alkylating agents in typical SN2 reactions. Look at the following electrostatic potential maps, and tell which is the better nucleophile, guanine or adenine. The reactive positions in each are indicated.
Mechanism Problems
Problem 28-16
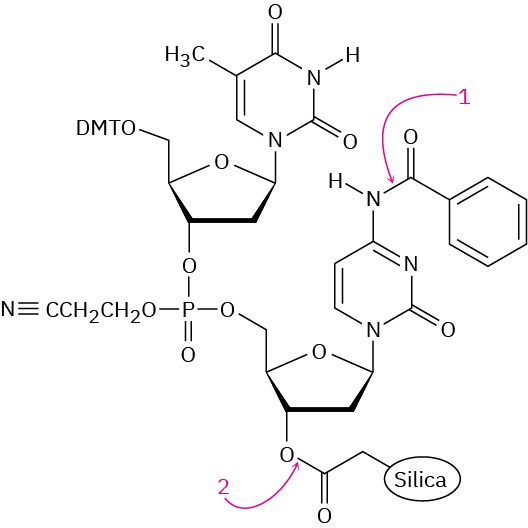
The final step in DNA synthesis is deprotection by treatment with aqueous ammonia. Show the mechanisms by which deprotection occurs at the points indicated in the following structure:
Problem 28-17
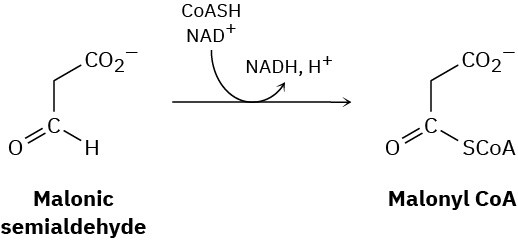
The final step in the metabolic degradation of uracil is the oxidation of malonic semialdehyde to give malonyl CoA. Propose a mechanism.
Problem 28-18
One of the steps in the biosynthesis of a nucleotide called inosine monophosphate is the formation of aminoimidazole ribonucleotide from formylglycinamidine ribonucleotide. Propose a mechanism.
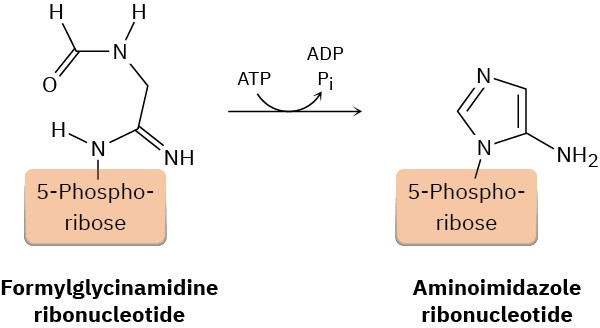
Problem 28-19
One of the steps in the metabolic degradation of guanine is hydrolysis to give xanthine. Propose a mechanism.
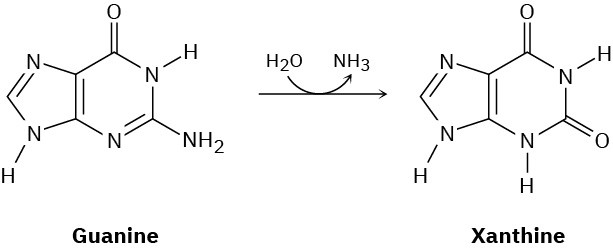
Problem 28-20
One of the steps in the biosynthesis of uridine monophosphate is the reaction of aspartate with carbamoyl phosphate to give carbamoyl aspartate followed by cyclization to form dihydroorotate. Propose mechanisms for both steps.
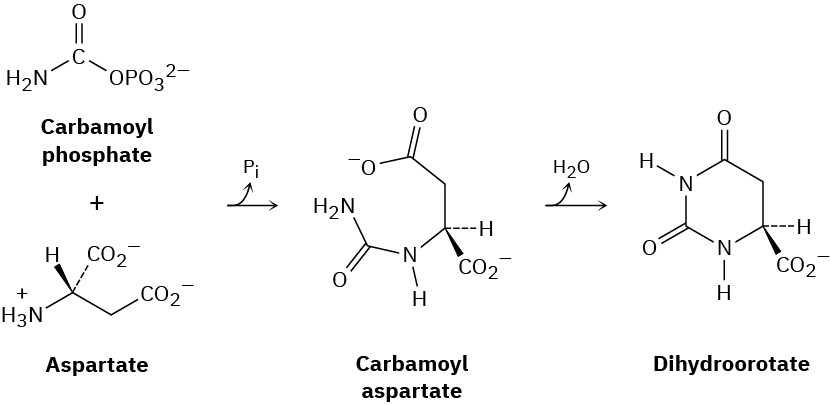
General Problems
Problem 28-21
Human brain natriuretic peptide (BNP) is a small peptide of 32 amino acids used in the treatment of congestive heart failure. How many nitrogen bases are present in the DNA that codes for BNP?
Problem 28-22
Human and horse insulin both have two polypeptide chains, with one chain containing 21 amino acids and the other containing 30 amino acids. They differ in primary structure at two places. At position 9 in one chain, human insulin has Ser and horse insulin has Gly; at position 30 in the other chain, human insulin has Thr and horse insulin has Ala. How must the DNA for the two insulins differ?
Problem 28-23
The DNA of sea urchins contains about 32% A. What percentages of the other three bases would you expect in sea urchin DNA? Explain.
Problem 28-24
The codon UAA stops protein synthesis. Why does the sequence UAA in the following stretch of mRNA not cause any problems?
-GCA-UUC-GAG-GUA-ACG-CCC-
Problem 28-25
Which of the following base sequences would most likely be recognized by a restriction endonuclease? Explain.
(a) GAATTC
(b) GATTACA
(c) CTCGAG
Problem 28-26
For what amino acids do the following ribonucleotide triplets code? (a)
AAU
(b) GAG
(c) UCC
(d) CAU
Problem 28-27
From what DNA sequences were each of the mRNA codons in Problem 28-26 transcribed? Problem 28-28
What anticodon sequences of tRNAs are coded for by the codons in Problem 28-26? Problem 28-29
Draw the complete structure of the ribonucleotide codon UAC. For what amino acid does this sequence code?
Problem 28-30
Draw the complete structure of the deoxyribonucleotide sequence from which the mRNA codon in Problem 28-29 was transcribed.
Problem 28-31
Give an mRNA sequence that will code for the synthesis of metenkephalin. Tyr-Gly-Gly-Phe-Met
Problem 28-32
Give an mRNA sequence that will code for the synthesis of angiotensin II. Asp-Arg-Val-Tyr-Ile-His-Pro-Phe
Problem 28-33
What amino acid sequence is coded for by the following DNA coding strand (sense strand)? (5′) CTT-CGA-CCA-GAC-AGC-TTT (3′)
Problem 28-34
What amino acid sequence is coded for by the following mRNA base sequence? (5′) CUA-GAC-CGU-UCC-AAG-UGA (3′)
Problem 28-35
If the DNA coding sequence -CAA-CCG-GAT- were miscopied during replication and became
-CGA-CCG-GAT-, what effect would there be on the sequence of the protein produced?
Problem 28-36
Show the steps involved in a laboratory synthesis of the DNA fragment with the sequence CTAG.
Problem 28-37
Draw the structure of cyclic adenosine monophosphate (cAMP), a messenger involved in the regulation of glucose production in the body. Cyclic AMP has a phosphate ring connecting the 3′- and 5′-hydroxyl groups on adenosine.
Problem 28-38
Valganciclovir, marketed as Valcyte, is an antiviral agent used for the treatment of cytomegalovirus. Called a prodrug, valganciclovir is inactive by itself but is rapidly converted in the intestine by hydrolysis of its ester bond to produce an active drug, called ganciclovir, along with an amino acid.
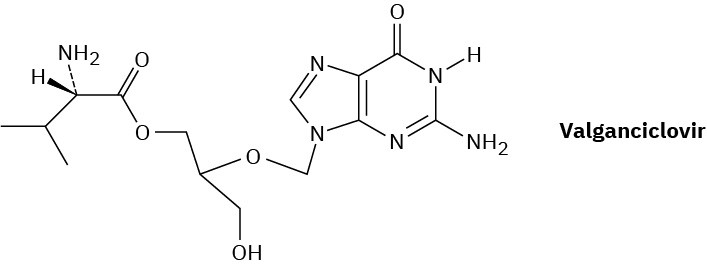
(a)
What amino acid is produced by hydrolysis of the ester bond in valganciclovir? (b)
What is the structure of ganciclovir? (c)
What atoms present in the nucleotide deoxyguanine are missing from ganciclovir? (d)
What role do the atoms missing from deoxyguanine play in DNA replication? (e)
How might valganciclovir interfere with DNA synthesis?

